The standard treatment of locally advanced esophageal cancer comprises multimodal treatment concepts including preoperative chemoradiotherapy (CRT) followed by radical surgical resection. However, despite intensified treatment approaches, 5-year survival rates are still low. Therefore, new strategies are required to overcome treatment resistance, and to improve patients’ outcome. In this study, we investigated the impact of Wnt/β-catenin signaling on CRT resistance in esophageal cancer cells. Experiments were conducted in adenocarcinoma and squamous cell carcinoma cell lines with varying expression levels of Wnt proteins and Wnt/β-catenin signaling activities. To investigate the effect of Wnt/β-catenin signaling on CRT responsiveness, we genetically or pharmacologically inhibited Wnt/β-catenin signaling. Our experiments revealed that inhibition of Wnt/β-catenin signaling sensitizes cell lines with robust pathway activity to CRT. In conclusion, Wnt/β-catenin activity may guide precision therapies in esophageal carcinoma patients.
- esophageal cancer
- chemoradiotherapy
- treatment resistance
- chemoradiotherapy-sensitization
- Wnt/β-catenin pathway
1. Introduction
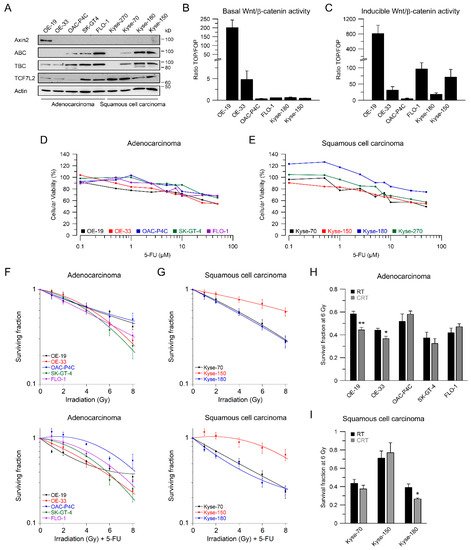
2. Esophageal Cancer Cell Lines Show Different Wnt/β-Catenin Pathway Activities
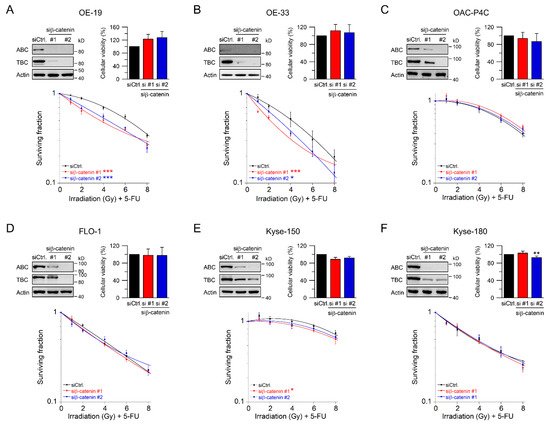
3. RNAi-Mediated Inhibition of Wnt/β-Catenin Signaling Sensitizes to CRT
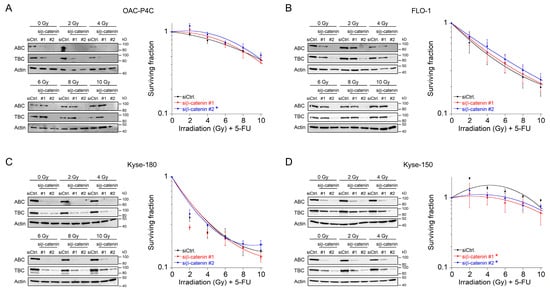
4. Fractionated Irradiation in Wnt/β-Catenin Signaling-Independent Cell Lines
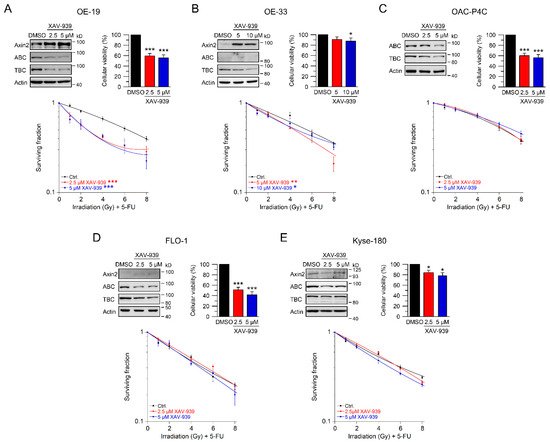
5. Using Small-Molecule Inhibitors as a Therapeutic Strategy to Sensitize Esophageal Cancer Cells
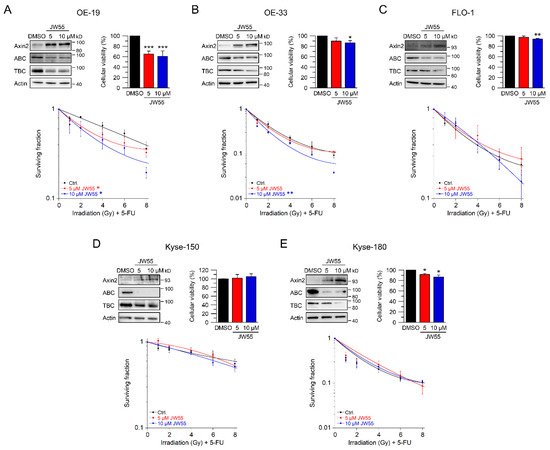
6. Discussion
Treatment resistance represents a fundamental problem in clinical oncology. In this context, esophageal cancer represents a prime example, because success rates remain low with dismal outcomes and 5-year survival rates ranging from 20% to 38% [1,7,9], indicating that for only a subset of patients, multimodal treatment concepts are effective [10,11].
In our set of esophageal cancer cell lines, we observed for both histological subtypes (SCC and AC) different response rates to RT and 5-FU-based CRT, measured by clonogenic survival. Comparing CRT responses of these cell lines (Figure 1F,G) to our observations in a panel of CRC cell lines [32], esophageal cancer cell lines appear to be more resistant to CRT. In our study, we showed that for a subset of cell lines with high basal and inducible Wnt/β-catenin signaling activity, sensitization to CRT upon inhibition of the pathway occurs. Inhibition of Wnt/β-catenin signaling was achieved by different methods (RNAi and pharmacological blockade), which resulted in re-sensitization of the resistant AC lines OE-19 and OE-33 to CRT. Since, for technical reasons, we were only able to assess basal activity in two SCC lines, both of which revealed no Wnt/β-catenin reporter activity, we currently cannot conclude the potential role of Wnt/β-catenin pathway inhibition in this subtype. However, other groups have investigated radiation resistance in SCC esophageal cancer cells, and provided evidence for a potential involvement of Wnt/β-catenin signaling in mediating RT resistance in this histological subtype [43–47]. In one study, the authors used an indirect approach to inhibit Wnt/β-catenin signaling via a microRNA, which influences the expression of Wnt/β-catenin-related genes. miRNA-381 was found to be downregulated in resistant SCC tissues and in cell lines exhibiting a re-sensitizing effect after expression, whereas inhibition thereof, promoted radiation resistance. However, this study was not primarily focused on Wnt/β-catenin signaling, it was rather a screen of microRNAs to compare tissues from primary esophageal SCC and recurrent esophageal SCC following RT [47]. In another study of esophageal SCC, two isogenic radioresistant cell lines were generated and showed changes in the expression levels of nuclear β-catenin and c-myc, which resulted in an enhanced RT resistance compared to the corresponding parental cells [48]. Mechanistically, it was demonstrated that Wnt/β-catenin signaling promotes DNA damage repair by transactivation of the high-mobility group box 1 protein (HMGB1) [48], an observation that is consistent with our investigation of radiation resistant CRC cells [24]. One limitation of this study is that the authors determined Wnt/β-catenin activity only by Western blot analysis and IF staining of β-catenin and c-myc, instead of measuring Wnt/β-catenin activity by TOPFlash/FOPFlash reporter assays, as standard in the field [27]. In this context, we observed that there was no correlation between β-catenin protein expression and basal Wnt/β-catenin activity, as shown for OE-19 and OE-33 cells. The molecular reasons are still unclear and demand further experimentation. Together, our data and these studies point to a potential role for inhibiting Wnt/β-catenin signaling as a therapeutic concept to increase responsiveness of esophageal cancer to CRT.
The potential relevance of Wnt/β-catenin signaling in mediating RT/CRT resistance has been demonstrated in other tumor entities, including CRC [23,24,49,50], prostate cancer [51], lung cancer [52], head and neck cancer [53], breast cancer and mammary gland cells [54], nasopharyngeal cancer [55], glioblastoma [56], and pancreatic cancer [57]. Although these reports underpin the relevance of Wnt/β-catenin signaling for radioresistance, the underlying mechanisms are still not fully understood. Wnt/β-catenin signaling triggers numerous cellular and molecular mechanisms presumably involved in drug efflux, DNA damage repair, inhibition of apoptosis, regulation of the cell cycle, cellular survival, reactive oxygen species (ROS), induction of epithelial to mesenchymal transition (EMT), and modification of the tumor microenvironment (TME) [25,26,58], which all can be connected to treatment resistance.
Until today, extensive efforts have been made in the development of small-molecule inhibitors that target the Wnt/β-catenin pathway, but none of them have yet reached clinical application as an FDA approved drug [34,35,50,59]. However, several inhibitors entered clinical testing, which include OMP-18R5 (vantictumab), a monoclonal antibody against Frizzled receptors, OMP-54F28, which binds to all Wnt-ligands, LGK974 and ETC-1922159 as examples for porcupine inhibitors preventing the production of bioactive Wnt-ligands [50]. Novel pharmacological concepts, which allow direct degradation of β-catenin, are under development [60]. Such data underscore the translational potential of our study.
From our data, only a subset of esophageal cancer cell lines showed basal Wnt/β-catenin activity and could be re-sensitized to CRT after pathway inhibition. Other pathways, such as IL-6/JAK/STAT signaling, were similarly shown to mediate CRT resistance in a subset of esophageal adenocarcinomas [61]. Here, multimodal stratification will help to tailor precise therapies for esophageal cancers. Although further studies will be needed to define esophageal cancers with high basal Wnt/β-catenin activity, they may then benefit from Wnt/β-catenin inhibitor-based CRT, our data point to a therapeutic concept with clinical potential.
7. Materials and Methods
7.1. Cell Lines and Cell Culture
Human esophageal cancer cell lines FLO-1, OAC-P4C, OE-19, OE-33, SK-GT-4 (all from adenocarcinoma), and Kyse-70, Kyse-150, Kyse-180, and Kyse-270 (all from squamous cell carcinoma) were obtained in 2013 directly from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (DSMZ, Braunschweig, Germany). The DSMZ ensures authenticity of these cell lines using short tandem repeat profiling [62]. Kyse cell lines were established by Shimada et al. [40]. After arrival, all cell lines were expanded and frozen down in aliquots. Cells were cultured in their recommended media (Invitrogen, Carlsbad, Germany), supplemented with 5% or 10% fetal bovine serum (Pan, Aidenbach, Germany), and 2 mM l-glutamine (BioWhittaker, Verviers, Belgium). For experimental use, cells older than 15 passages were discarded. Periodically, mycoplasma contamination was excluded using the MycoAlert® Mycoplasma Detection Kit (Lonza, Cologne, Germany).
7.2. Western Blot Analysis
Western blot analysis was performed as previously described [23,24,63]. Briefly, cells were lysed in NP-40 whole cell lysis buffer, and 20 µg of protein was loaded and resolved on a 10% bis-tris polyacrylamide gel. Protein transfer was performed by semi-dry blotting onto a polyvinylidene difluoride membrane (PVDF, GE Healthcare, Little Chalfont, UK), followed by antibody incubation and detection by the ImageQuant LAS 4000 mini CCD camera system (GE Healthcare). Table S2 includes the corresponding antibodies and experimental conditions. Original blot images and calculated band intensities (ImageJ software, version 1.52a, Wayne Rasband, National Institutes of Health, Bethesda, ) are provided in Figure S2.
7.3. Dual Luciferase Reporter Assay
Plasmid transfections were performed as described before [23]. Briefly, for determination of basal Wnt/β-catenin activity, cells were transfected with the reporter plasmids SuperTOPFlash, SuperFOPFlash (TOP: #12456, FOP: #12457, Addgene, Cambridge, MA), and Renilla (Promega, Madison, WI) using X-tremeGENE HP DNA Transfection Reagent (Roche, Penzberg, Germany). To measure the inducibility of the pathway, mutated pCl-neo-β-catenin-S33Y was co-transfected. pCI-neo-β-catenin-S33Y was a gift from Bert Vogelstein (Addgene plasmid # 16519, http://n2t.net/addgene:16519, accessed on 16. 08. 2021). Twenty-four hours after transfection, cells were lysed by passive lysis buffer (Promega), and both firefly and Renilla luciferase activity was measured in a microplate reader (Mithras LB940, Berthold Technologies, Bad Wildbad, Germany). Relative basal transcriptional activation was calculated by dividing Renilla-normalized values of SuperTOPFlash and SuperFOPFlash, whereas inducible activity was calculated by dividing samples that were co-transfected with pCl-neo-β-catenin-S33Y. Detailed experimental conditions are shown in Table S3.
7.4. Cellular Viability Assay
Cellular viability following 5-FU treatment, synthetic small interfering RNA (siRNA)-transfections, or inhibitor treatment (XAV-939, JW55) was assessed using the CellTiter-Blue® reagent (Promega). Reduction of resazurin to resorufin was measured at various time points after the respective treatment using a plate reader (VICTOR™ X4, Perkin Elmer, Waltham, MA, USA) according to the manufacturer’s instructions. Cellular viability of treated or siRNA-transfected cells was compared to untreated cells, or cells transfected with a non-silencing control siRNA (siCtrl.). Detailed information can be found in Table S3.
7.5. siRNA Transfection
Transfections with siRNA duplexes were performed as previously described [63]. Briefly, for cellular viability assays, cells were reverse transfected with siRNA (Qiagen, Hilden, Germany; Dharmacon/Thermo Fisher Scientific, Schwerte, Germany) using RNAiMAX (Invitrogen) or HiPerFect (Qiagen). For colony formation assays and Western blot analyses, cells were transfected using nucleofector technology (Lonza). Additional information about transfection conditions and siRNA sequences can be found in Tables S3–S5.
7.6. Chemoradiotherapy and Colony Formation Assays
To test the sensitivity to CRT, standard CFA were conducted as previously described [23,24,63]. Briefly, tumor cells growing in log-phase were seeded as single-cell suspensions into six-well plates. Eight hours after seeding, cells were treated by 3 µM 5-FU (Sigma-Aldrich, Steinheim, Germany), incubated overnight, and subsequently irradiated with single doses of 1, 2, 4, 6, and 8 Gy of X-rays (Gulmay Medical, Camberley, UK). To test the influence of different treatments, cells were either transfected with siRNA, or exposed to inhibitors before irradiation. For irradiation experiments in a fractionated setting, cells were repeatedly irradiated with 2 Gy every 12 h, until a total dose of 10 Gy was reached (Figure S1). After colony formation in the control wells, cells were fixed with 70% ethanol, stained with Mayer’s hemalum solution (Merck KGaA, Darmstadt, Germany), and counted. Colonies were analyzed according to Franken et al. [29]. For a comprehensive evaluation of the effects of the respective treatments (siRNA, inhibitors), a radiation enhancement ratio (RER) was calculated to illustrate the magnitude of radiation sensitization. The RER is defined as the ratio of survival fractions (SF) without and with treatments for a specific dose [39,64]. All experiments were performed in technical triplicates, and independently repeated at least three times (biological replicates). Table S5 shows all experimental conditions for irradiation experiments.
7.7. Statistical Analysis
Statistical analyses of SF6 levels for RT and CRT, cellular viability, and luciferase reporter activity experiments were performed using an unpaired two-tailed Student’s t-test in Microsoft Excel and visualized in Grapher (version 8.2.460). p-values < 0.05 were scored as significant. For analyses of the irradiation data, analysis of variance (ANOVA) was used to calculate significant differences between the control group and treatment group. All analyses were performed using Microsoft Excel software Add-in “Data Analysis” (ANOVA: Two-Factor with Replication). For visualization, irradiation data are presented as mean and standard error of the mean (SEM) from at least three independent experiments using the software KaleidaGraph (version 4.1.0). Again, p-values < 0.05 were considered significant, suggesting an influence of the treatment on the dose response. All p-values determined in this study are provided in Table S1.
8. References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33, doi:10.3322/caac.21654.
- Kelly, R.J. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer. J. Compr. Cancer Netw. JNCCN 2019, 17, 1009–1014, doi:10.6004/jnccn.2019.7337.
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal cancer. Lancet (London, England) 2017, 390, 2383–2396, doi:10.1016/s0140-6736(17)31462-9.
- Al-Batran, S.E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet (London, England) 2019, 393, 1948–1957, doi:10.1016/s0140-6736(18)32557-1.
- Allum, W.H.; Stenning, S.P.; Bancewicz, J.; Clark, P.I.; Langley, R.E. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5062–5067, doi:10.1200/jco.2009.22.2083.
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van de Velde, C.J.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. Engl. J. Med. 2006, 355, 11–20, doi:10.1056/NEJMoa055531.
- Shapiro, J.; van Lanschot, J.J.B.; Hulshof, M.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015, 16, 1090–1098, doi:10.1016/s1470-2045(15)00040-6.
- van Hagen, P.; Hulshof, M.C.; van Lanschot, J.J.; Steyerberg, E.W.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.; Richel, D.J.; Nieuwenhuijzen, G.A.; Hospers, G.A.; Bonenkamp, J.J.; et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. Engl. J. Med. 2012, 366, 2074–2084, doi:10.1056/NEJMoa1112088.
- Eyck, B.M.; van Lanschot, J.J.B.; Hulshof, M.; van der Wilk, B.J.; Shapiro, J.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; et al. Ten-Year Outcome of Neoadjuvant Chemoradiotherapy Plus Surgery for Esophageal Cancer: The Randomized Controlled CROSS Trial. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 1995–2004, doi:10.1200/jco.20.03614.
- Davies, A.R.; Myoteri, D.; Zylstra, J.; Baker, C.R.; Wulaningsih, W.; Van Hemelrijck, M.; Maisey, N.; Allum, W.H.; Smyth, E.; Gossage, J.A.; et al. Lymph node regression and survival following neoadjuvant chemotherapy in oesophageal adenocarcinoma. J. Surg. 2018, 105, 1639–1649, doi:10.1002/bjs.10900.
- Noble, F.; Lloyd, M.A.; Turkington, R.; Griffiths, E.; O'Donovan, M.; O'Neill, J.R.; Mercer, S.; Parsons, S.L.; Fitzgerald, R.C.; Underwood, T.J. Multicentre cohort study to define and validate pathological assessment of response to neoadjuvant therapy in oesophagogastric adenocarcinoma. J. Surg. 2017, 104, 1816–1828, doi:10.1002/bjs.10627.
- Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175, doi:10.1038/nature20805
- Ma, F.; Laster, K.; Nie, W.; Liu, F.; Kim, D.J.; Lee, M.H.; Bai, R.; Yang, R.; Liu, K.; Dong, Z. Heterogeneity Analysis of Esophageal Squamous Cell Carcinoma in Cell Lines, Tumor Tissues and Patient-Derived Xenografts. Cancer 2021, 12, 3930–3944, doi:10.7150/jca.52286.
- Walker, R.C.; Underwood, T.J. Molecular pathways in the development and treatment of oesophageal cancer. Best Pract. Res. Clin. Gastroenterol. 2018, -37, 9–15, doi:10.1016/j.bpg.2018.11.013.
- Chiu, W.C.; Lee, Y.C.; Su, Y.H.; Wang, Y.Y.; Tsai, C.H.; Hou, Y.A.; Wang, C.H.; Huang, Y.F.; Huang, C.J.; Chou, S.H.; et al. The Synthetic β-Nitrostyrene Derivative CYT-Rx20 Inhibits Esophageal Tumor Growth and Metastasis via PI3K/AKT and STAT3 Pathways. PLoS ONE 2016, 11, e0166453, doi:10.1371/journal.pone.0166453.
- Keld, R.R.; Ang, Y.S. Targeting key signalling pathways in oesophageal adenocarcinoma: A reality for personalised medicine? World J. Gastroenterol. 2011, 17, 2781–2790, doi:10.3748/wjg.v17.i23.2781.
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet (London, England) 2010, 376, 687–697, doi:10.1016/s0140-6736(10)61121-x.
- Clément, G.; Braunschweig, R.; Pasquier, N.; Bosman, F.T.; Benhattar, J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett's esophagus. Oncogene 2006, 25, 3084–3092, doi:10.1038/sj.onc.1209338.
- Clément, G.; Jablons, D.M.; Benhattar, J. Targeting the Wnt signaling pathway to treat Barrett's esophagus. Expert Opin. Ther. Targets 2007, 11, 375–389, doi:10.1517/14728222.11.3.375.
- Denlinger, C.E.; Thompson, R.K. Molecular basis of esophageal cancer development and progression. Clin. North Am. 2012, 92, 1089–1103, doi:10.1016/j.suc.2012.07.002.
- Kalatskaya, I. Overview of major molecular alterations during progression from Barrett's esophagus to esophageal adenocarcinoma. N. Y. Acad. Sci. 2016, 1381, 74–91, doi:10.1111/nyas.13134.
- Moyes, L.H.; McEwan, H.; Radulescu, S.; Pawlikowski, J.; Lamm, C.G.; Nixon, C.; Sansom, O.J.; Going, J.J.; Fullarton, G.M.; Adams, P.D. Activation of Wnt signalling promotes development of dysplasia in Barrett's oesophagus. Pathol. 2012, 228, 99–112, doi:10.1002/path.4058.
- Emons, G.; Spitzner, M.; Reineke, S.; Moller, J.; Auslander, N.; Kramer, F.; Hu, Y.; Beissbarth, T.; Wolff, H.A.; Rave-Frank, M.; et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/beta-catenin Signaling. Mol. Cancer Res. 2017, 15, 1481–1490, doi:10.1158/1541-7786.MCR-17-0205.
- Kendziorra, E.; Ahlborn, K.; Spitzner, M.; Rave-Fränk, M.; Emons, G.; Gaedcke, J.; Kramer, F.; Wolff, H.A.; Becker, H.; Beissbarth, T.; et al. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011, 32, 1824–1831, doi:10.1093/carcin/bgr222.
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Cell Dev. Biol. 2021, 9, 650772, doi:10.3389/fcell.2021.650772.
- Zhao, Y.; Tao, L.; Yi, J.; Song, H.; Chen, L. The Role of Canonical Wnt Signaling in Regulating Radioresistance. Cell Physiol. Biochem. Int. J. Exp. Cell Physiol. Biochem. Pharmacol. 2018, 48, 419–432, doi:10.1159/000491774.
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/-colon carcinoma. Science (New York, N.Y.) 1997, 275, 1784–1787, doi:10.1126/science.275.5307.1784.
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science (New York, N.Y.) 1997, 275, 1787–1790, doi:10.1126/science.275.5307.1787.
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Protoc. 2006, 1, 2315–2319, doi:10.1038/nprot.2006.339.
- Carpentier, Y.; Demange, L.; Loirette, M.; Hivet, J.; Desoize, B. Chronology of combined chemotherapy (5FU) and radiotherapy. I. In vitro study. Res. 1993, 13, 2177–2180.
- Seiwert, T.Y.; Salama, J.K.; Vokes, E.E. The concurrent chemoradiation paradigm—general principles. Clin. Practice Oncol. 2007, 4, 86–100, doi:10.1038/ncponc0714.
- Spitzner, M.; Emons, G.; Kramer, F.; Gaedcke, J.; Rave-Fränk, M.; Scharf, J.G.; Burfeind, P.; Becker, H.; Beissbarth, T.; Ghadimi, B.M.; et al. A gene expression signature for chemoradiosensitivity of colorectal cancer cells. J. Radiat. Oncol. Biol. Phys. 2010, 78, 1184–1192, doi:10.1016/j.ijrobp.2010.06.023.
- Franken, N.A.; Oei, A.L.; Kok, H.P.; Rodermond, H.M.; Sminia, P.; Crezee, J.; Stalpers, L.J.; Barendsen, G.W. Cell survival and radiosensitisation: Modulation of the linear and quadratic parameters of the LQ model (Review). J. Oncol. 2013, 42, 1501–1515, doi:10.3892/ijo.2013.1857.
- Chatterjee, A.; Paul, S.; Bisht, B.; Bhattacharya, S.; Sivasubramaniam, S.; Paul, M.K. Advances in targeting the WNT/β-catenin signaling pathway in cancer. Drug Discov. Today , Jul 10:S1359-6446(21)00317-2, doi:10.1016/j.drudis.2021.07.007.
- Jackstadt, R.; Hodder, M.C.; Sansom, O.J. WNT and beta-Catenin in Cancer: Genes and Therapy. Rev. Cancer Biol. 2020, 4, 177–196, doi:10.1146/annurev-cancerbio-030419-033628.
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science (New York, N.Y.) 2014, , (6205):1248012, doi:10.1126/science.1248012.
- Nusse, R. Wnt signaling. Cold Spring Harb. Perspect. Biol. 2012, , (5):a011163, doi:10.1101/cshperspect.a011163.
- Paireder, M.; Jomrich, G.; Kristo, I.; Asari, R.; Rieder, E.; Beer, A.; Ilhan-Mutlu, A.; Preusser, M.; Schmid, R.; Schoppmann, S.F. Modification of preoperative radiochemotherapy for esophageal cancer (CROSS protocol) is safe and efficient with no impact on surgical morbidity. . 2020, 196, 779–786, doi:10.1007/s00066-020-01594-z.
- Subiel, A.; Ashmore, R.; Schettino, G. Standards and Methodologies for Characterizing Radiobiological Impact of High-Z Nanoparticles. Theranostics 2016, 6, 1651–1671, doi:10.7150/thno.15019.
- Shimada, Y.; Imamura, M.; Wagata, T.; Yamaguchi, N.; Tobe, T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992, 69, 277–284, doi:10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c.
- Huang, S.M.; Mishina, Y.M.; Liu, S.; Cheung, A.; Stegmeier, F.; Michaud, G.A.; Charlat, O.; Wiellette, E.; Zhang, Y.; Wiessner, S.; et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009, 461, 614–620, doi:10.1038/nature08356.
- Waaler, J.; Machon, O.; Tumova, L.; Dinh, H.; Korinek, V.; Wilson, S.R.; Paulsen, J.E.; Pedersen, N.M.; Eide, T.J.; Machonova, O.; et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012, 72, 2822–2832, doi:10.1158/0008-5472.Can-11-3336.
- Su, H.; Jin, X.; Zhang, X.; Xue, S.; Deng, X.; Shen, L.; Fang, Y.; Xie, C. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol. Int. 2014, 38, 318–325, doi:10.1002/cbin.10202.
- Su, H.; Wu, Y.; Fang, Y.; Shen, L.; Fei, Z.; Xie, C.; Chen, M. MicroRNA‑301a targets WNT1 to suppress cell proliferation and migration and enhance radiosensitivity in esophageal cancer cells. Rep. 2019, 41, 599–607, doi:10.3892/or.2018.6799.
- Su, H.; Wu, Y.; Fang, Y.; Shen, L.; Fei, Z.; Xie, C.; Chen, M. MicroRNA‑301a targets WNT1 to suppress cell proliferation and migration and enhance radiosensitivity in esophageal cancer cells. Rep. 2019, 41, 599–607, doi:10.3892/or.2018.6799.
- Zhou, H.; Zhang, G.; Xue, X.; Yang, Y.; Yang, Y.; Chang, X.; Ge, X.; Xiao, Z.; Guo, H.; Wang, Y. Identification of novel NRAGE involved in the radioresistance of esophageal cancer cells. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 8741–8752, doi:10.1007/s13277-015-4747-6.
- Zhou, S.; Ye, W.; Ren, J.; Shao, Q.; Qi, Y.; Liang, J.; Zhang, M. MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. J. Cancer Res. 2015, 5, 267–277.
- Zhao, Y.; Yi, J.; Tao, L.; Huang, G.; Chu, X.; Song, H.; Chen, L. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis. 2018, 9, 433, doi:10.1038/s41419-018-0466-4.
- Kleeman, S.O.; Koelzer, V.H.; Jones, H.J.; Vazquez, E.G.; Davis, H.; East, J.E.; Arnold, R.; Koppens, M.A.; Blake, A.; Domingo, E.; et al. Exploiting differential Wnt target gene expression to generate a molecular biomarker for colorectal cancer stratification. Gut 2020, 69, 1092–1103, doi:10.1136/gutjnl-2019-319126.
- Wang, Z.; Li, Z.; Ji, H. Direct targeting of β-catenin in the Wnt signaling pathway: Current progress and perspectives. Res. Rev. 2021, 41, 2109–2129, doi:10.1002/med.21787.
- He, Z.; Shen, F.; Qi, P.; Zhai, Z.; Wang, Z. miR-541-3p enhances the radiosensitivity of prostate cancer cells by inhibiting HSP27 expression and downregulating β-catenin. Cell Death Discov. 2021, 7, 18, doi:10.1038/s41420-020-00387-8.
- Zhang, Q.; Gao, M.; Luo, G.; Han, X.; Bao, W.; Cheng, Y.; Tian, W.; Yan, M.; Yang, G.; An, J. Enhancement of Radiation Sensitivity in Lung Cancer Cells by a Novel Small Molecule Inhibitor That Targets the β-Catenin/Tcf4 Interaction. PLoS ONE 2016, 11, e0152407, doi:10.1371/journal.pone.0152407.
- Chang, H.W.; Roh, J.L.; Jeong, E.J.; Lee, S.W.; Kim, S.W.; Choi, S.H.; Park, S.K.; Kim, S.Y. Wnt signaling controls radiosensitivity via cyclooxygenase-2-mediated Ku expression in head and neck cancer. J. Cancer 2008, 122, 100–107, doi:10.1002/ijc.23069.
- Woodward, W.A.; Chen, M.S.; Behbod, F.; Alfaro, M.P.; Buchholz, T.A.; Rosen, J.M. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Natl. Acad. Sci. USA 2007, 104, 618–623, doi:10.1073/pnas.0606599104.
- Li, G.; Liu, Y.; Su, Z.; Ren, S.; Zhu, G.; Tian, Y.; Qiu, Y. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. J. Cancer (Oxford, England: 1990) 2013, 49, 2596–2607, doi:10.1016/j.ejca.2013.03.001.
- Kim, Y.; Kim, K.H.; Lee, J.; Lee, Y.A.; Kim, M.; Lee, S.J.; Park, K.; Yang, H.; Jin, J.; Joo, K.M.; et al. Wnt activation is implicated in glioblastoma radioresistance. Investig. J. Tech. Methods Pathol. 2012, 92, 466–473, doi:10.1038/labinvest.2011.161.
- Watson, R.L.; Spalding, A.C.; Zielske, S.P.; Morgan, M.; Kim, A.C.; Bommer, G.T.; Eldar-Finkelman, H.; Giordano, T.; Fearon, E.R.; Hammer, G.D.; et al. GSK3beta and beta-catenin modulate radiation cytotoxicity in pancreatic cancer. Neoplasia (New York, N.Y.) 2010, 12, 357–365, doi:10.1593/neo.92112.
- Yang, Y.; Zhou, H.; Zhang, G.; Xue, X. Targeting the canonical Wnt/β-catenin pathway in cancer radioresistance: Updates on the molecular mechanisms. J. Cancer Res. Ther. 2019, 15, 272–277, doi:10.4103/jcrt.JCRT_421_18.
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999, doi:10.1016/j.cell.2017.05.016.
- Liao, H.; Li, X.; Zhao, L.; Wang, Y.; Wang, X.; Wu, Y.; Zhou, X.; Fu, W.; Liu, L.; Hu, H.G.; et al. A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell Discov. 2020, 6, 35, doi:10.1038/s41421-020-0171-1.
- Ebbing, E.A.; van der Zalm, A.P.; Steins, A.; Creemers, A.; Hermsen, S.; Rentenaar, R.; Klein, M.; Waasdorp, C.; Hooijer, G.K.J.; Meijer, S.L.; et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Natl. Acad. Sci. USA 2019, 116, 2237–2242, doi:10.1073/pnas.1820459116.
- Masters, J.R.; Thomson, J.A.; Daly-Burns, B.; Reid, Y.A.; Dirks, W.G.; Packer, P.; Toji, L.H.; Ohno, T.; Tanabe, H.; Arlett, C.F.; et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Natl. Acad. Sci. USA 2001, 98, 8012–8017, doi:10.1073/pnas.121616198.
- Spitzner, M.; Roesler, B.; Bielfeld, C.; Emons, G.; Gaedcke, J.; Wolff, H.A.; Rave-Fränk, M.; Kramer, F.; Beissbarth, T.; Kitz, J.; et al. STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. J. Cancer 2014, 134, 997–1007, doi:10.1002/ijc.28429.
- Zeng, L.; Boggs, D.H.; Xing, C.; Zhang, Z.; Anderson, J.C.; Wajapeyee, N.; Veale, C.; Bredel, M.; Shi, L.Z.; Bonner, J.A.; et al. Combining PARP and DNA-PK Inhibitors With Irradiation Inhibits HPV-Negative Head and Neck Cancer Squamous Carcinoma Growth. Genet. 2020, 11, 1036, doi:10.3389/fgene.2020.01036.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33.
- Kelly, R.J. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 1009–1014.
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal cancer. Lancet 2017, 390, 2383–2396.
- Al-Batran, S.E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957.
- Allum, W.H.; Stenning, S.P.; Bancewicz, J.; Clark, P.I.; Langley, R.E. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5062–5067.
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van de Velde, C.J.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 2006, 355, 11–20.
- Shapiro, J.; van Lanschot, J.J.B.; Hulshof, M.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015, 16, 1090–1098.
- van Hagen, P.; Hulshof, M.C.; van Lanschot, J.J.; Steyerberg, E.W.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.; Richel, D.J.; Nieuwenhuijzen, G.A.; Hospers, G.A.; Bonenkamp, J.J.; et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 2012, 366, 2074–2084.
- Eyck, B.M.; van Lanschot, J.J.B.; Hulshof, M.; van der Wilk, B.J.; Shapiro, J.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; et al. Ten-Year Outcome of Neoadjuvant Chemoradiotherapy Plus Surgery for Esophageal Cancer: The Randomized Controlled CROSS Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 1995–2004.
- Davies, A.R.; Myoteri, D.; Zylstra, J.; Baker, C.R.; Wulaningsih, W.; Van Hemelrijck, M.; Maisey, N.; Allum, W.H.; Smyth, E.; Gossage, J.A.; et al. Lymph node regression and survival following neoadjuvant chemotherapy in oesophageal adenocarcinoma. Br. J. Surg. 2018, 105, 1639–1649.
- Noble, F.; Lloyd, M.A.; Turkington, R.; Griffiths, E.; O’Donovan, M.; O’Neill, J.R.; Mercer, S.; Parsons, S.L.; Fitzgerald, R.C.; Underwood, T.J. Multicentre cohort study to define and validate pathological assessment of response to neoadjuvant therapy in oesophagogastric adenocarcinoma. Br. J. Surg. 2017, 104, 1816–1828.
- Emons, G.; Spitzner, M.; Reineke, S.; Moller, J.; Auslander, N.; Kramer, F.; Hu, Y.; Beissbarth, T.; Wolff, H.A.; Rave-Frank, M.; et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/beta-catenin Signaling. Mol. Cancer Res. 2017, 15, 1481–1490.
- Kendziorra, E.; Ahlborn, K.; Spitzner, M.; Rave-Fränk, M.; Emons, G.; Gaedcke, J.; Kramer, F.; Wolff, H.A.; Becker, H.; Beissbarth, T.; et al. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011, 32, 1824–1831.
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Front. Cell Dev. Biol. 2021, 9, 650772.
- Zhao, Y.; Tao, L.; Yi, J.; Song, H.; Chen, L. The Role of Canonical Wnt Signaling in Regulating Radioresistance. Cell Physiol. Biochem. Int. J. Exp. Cell Physiol. Biochem. Pharmacol. 2018, 48, 419–432.
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/-colon carcinoma. Science 1997, 275, 1784–1787.
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997, 275, 1787–1790.
- Chatterjee, A.; Paul, S.; Bisht, B.; Bhattacharya, S.; Sivasubramaniam, S.; Paul, M.K. Advances in targeting the WNT/β-catenin signaling pathway in cancer. Drug Discov. Today 2021.
- Jackstadt, R.; Hodder, M.C.; Sansom, O.J. WNT and beta-Catenin in Cancer: Genes and Therapy. Annu. Rev. Cancer Biol. 2020, 4, 177–196.
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 6205–1248012.
- Nusse, R. Wnt signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011163.
- Paireder, M.; Jomrich, G.; Kristo, I.; Asari, R.; Rieder, E.; Beer, A.; Ilhan-Mutlu, A.; Preusser, M.; Schmid, R.; Schoppmann, S.F. Modification of preoperative radiochemotherapy for esophageal cancer (CROSS protocol) is safe and efficient with no impact on surgical morbidity. Strahlenther. Onkol. 2020, 196, 779–786.
- Subiel, A.; Ashmore, R.; Schettino, G. Standards and Methodologies for Characterizing Radiobiological Impact of High-Z Nanoparticles. Theranostics 2016, 6, 1651–1671.
- Shimada, Y.; Imamura, M.; Wagata, T.; Yamaguchi, N.; Tobe, T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992, 69, 277–284.
- Huang, S.M.; Mishina, Y.M.; Liu, S.; Cheung, A.; Stegmeier, F.; Michaud, G.A.; Charlat, O.; Wiellette, E.; Zhang, Y.; Wiessner, S.; et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009, 461, 614–620.
- Waaler, J.; Machon, O.; Tumova, L.; Dinh, H.; Korinek, V.; Wilson, S.R.; Paulsen, J.E.; Pedersen, N.M.; Eide, T.J.; Machonova, O.; et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012, 72, 2822–2832.
