Small noncoding RNAs (sRNA) appear to play a key role in extracellular vesicle (EV)-mediated information transfer. Within the vesicular envelope, RNAs are well protected from degradation and can be shuttled between individuals from one and the same species and beyond. Various communication routes have been discovered such as mother-infant-interaction via breast milk, diverse host-pathogen-relations, and dietary uptake of food derived EVs, proving that EV-mediated inter-kingdom regulation is more than a random event.
- extracellular vesicles
- cross-kingdom RNA interference
- host-pathogen-interaction
1. Introduction
After the observation that EV-mediated information transfer is not limited to one organism, species or kingdoms, the one central question of EV research became, “Why can EVs overcome kingdom boundaries?” Investigations on EV-mediated regulation processes, from mother–infant to host–pathogen interaction, might elucidate this query (Table 1).
Table 1.
EV mediated regulation processes.
|
EV Mediated Regulations |
Examples |
References |
|
|
Inter-individual regulation |
mother ↔ foetus |
[169] |
|
|
mother → infant regulation |
[28,170,171] |
||
|
elevated fungal virulence |
[172,173] |
||
|
elevated bacterial virulence/drug resistance within the same species |
[148,153,156,174] |
||
|
Interspecies regulation |
dietary uptake, e.g., bovine milk → other mammals |
[175,176] |
|
|
pathogen-host interactions, e.g., helminth ↔ animal host |
[26,177] |
||
|
elevated bacterial virulence/drug resistance beyond species boundaries |
[148,153,156,174,178] |
||
|
archaeal antimicrobial proteins inhibit growth of other archaea |
[12,165] |
||
|
archaeal DNA tranfer |
[163,168] |
||
|
Inter-kingdom regulation |
pathogen–host interactions: |
plant ↔ fungus |
[27,91,97,179–181] |
|
animal ↔ fungus |
[119,124,172] |
||
|
bacteria ↔ animal |
[156,174,182–188] |
||
|
dietary uptake, e.g., rice → mammal |
[18,189] |
||
|
archaeal antimicrobial proteins inhibit bacterial growth |
[12,165] |
||
|
EV Mediated Regulations |
Examples |
References |
|
|
Inter-individual regulation |
mother ↔ foetus |
[1] |
|
|
mother → infant regulation |
|||
|
elevated fungal virulence |
|||
|
elevated bacterial virulence/drug resistance within the same species |
|||
|
Interspecies regulation |
dietary uptake, e.g., bovine milk → other mammals |
||
|
pathogen-host interactions, e.g., helminth ↔ animal host |
|||
|
elevated bacterial virulence/drug resistance beyond species boundaries |
|||
|
archaeal antimicrobial proteins inhibit growth of other archaea |
|||
|
archaeal DNA tranfer |
|||
|
Inter-kingdom regulation |
pathogen–host interactions: |
plant ↔ fungus |
|
|
animal ↔ fungus |
|||
|
bacteria ↔ animal |
[9][10][27][28][29][30][31][32][33] |
||
|
dietary uptake, e.g., rice → mammal |
|||
|
archaeal antimicrobial proteins inhibit bacterial growth |
|||
2. Inter-Individual, Interspecies, and Inter-Kingdom Regulation
In addition to mitogenic lipids and signaling proteins, sRNAs are considered to be crucial regulatory elements in EV-mediated (inter-kingdom) communication[36] [99]. They are able to manipulate various biological processes, such as cell growth, differentiation, development, metabolism, and apoptosis[37][38] [19,20]. Stability and absorption of sRNA are obviously critical aspects of bioavailability for recipient organisms or cells. In contrast to traditional persuasions on the stability of extracellular RNA, a few studies have shown surprisingly high pH-, temperature-, and RNase-resistances for sRNA in mammalian body fluids[13][39][40][41][42][43] [26,190–194], as well as for plant sRNAs[44] [45][46][47][48][21,80,96,195,196]. The vesicular envelope of EVs is thought to be decisive for the enhanced sRNA stability. This assumption is strongly underlined by the fact that severe losses of sRNA are detectable after pasteurization and homogenization or after ultrasonic exosome depletion of bovine milk[2][49][50] [28,175,197]. Furthermore, the envelope also provides a vehicle for cellular uptake of the cargo, not only in the intestine[2][49][12][41][51][52][53] [28,175,176,192,198–200].
Since EVs have been found in the milk of distinct mammals, such as pork, cow, or human, increasing numbers of inter-individual and interspecies regulation processes are being assumed highly probable[2][41][54][55][56][57] [28,192,201–204]. Moreover, increased serum levels of bovine milk specific sRNA were detected in humans after consumption of cow´s milk[11] [175]. Until today, we are lacking reliable studies on physiological or pathological effects of ingested EVs on humans, while a broad range of such effects is conceivable. This assumption is supported by investigations that have shown that a breastfed infant profits from ingested milk-derived sRNAs by elevated T-cell levels and enhanced differentiation of B cells[38][2][41][54] [20,28,192,201].
Although there has been previous evidence for inter-kingdom regulation mediated by sRNAs[58][59][60][61] [205–208], the study by Zhang et al., 2012 was somehow paradigm shifting. Their finding, that the dietary uptake of a particular plant-derived micro RNA can measurably affect the metabolism of a mammal[62] [189], quickly ignited increased interest in this field.
Probably, fungal cells send EVs in order to downregulate host immune response. Observations in both human–fungus and plant–fungus interactions suggest fungal virulence to be strongly enhanced by inter-kingdom RNA interference, enabled by sRNA containing EVs[26][5][22][24][63] [124,172,179,181,209]. Conversely, plants send sRNA to silence fungal virulence genes, which has recently also been related to EVs[19][20][21][64][63][65] [27,91,97,106,210,211].
In the area of difficult-to-treat infections, OMVs play a major role in drug resistance because they transfer resistance genes (DNA) between bacteria, even of different origin[7] [148]. Many OMVs from pathogenic bacteria were found to have surface proteins, which can readily interact with mammalian host cells. These interaction mechanisms make OMVs a pivotal element of trans-kingdom and host-cell communication by letting them interact in a highly specific manner[66] [212]. OMVs have been shown to carry PAMPs, including lipopolysaccharides, and can transfer other virulence associated factors[67] [213]. These factors can trigger strong immune responses in host cells, while OMVs act as immunomodulators, for example, by leading to expression of receptors on macrophages to specifically recognize the pathogen[68] [214]. As OMVs can help pathogenic bacteria to persist attack by the mammalian immune system, they strongly contribute to the cause of infectious disease[10][27] [174,182]. Prokaryotic pathogens such as Bacillus anthracis Cohn[28] [183], Helicobacter pylori (Marshall) Goodwin[29] [184], Neisseria gonorrhoeae (Zopf) Trevisan[30] [185], Pseudomonas aeruginosa (Schroeter) Migula[31] [186], and Streptococcus pneumoniae (Klein) Chester[32] [187], as well as eukarytotic pathogens such as Leishmania spp. Ross[69] [215], Plasmodium spp. Marchiafava et Celli[70] [216], and Trichomonas vaginalis Donné[71] [217] similarly send EVs to increase their contagiousness[33][72][73][74] [188,218–220]. This phenomenon is not limited to unicellular organisms, since helminths also modulate host immunity, as Heligosomoides polygyrus Dujardin[13] [26] and Dicrocoelium dendriticum Rudolphi[75] [177].
Overall, EVs appear to be potent agents in regulation processes, crossing not only the borders of species but rather of kingdoms or even empires. Therefore, they enhance an arms race in host–pathogen interaction[64][23] [106,180]. But do exosomes also facilitate intercellular communication beyond the animal kingdom? Especially host–pathogen interactions imply the possibility of host-host and pathogen-pathogen signaling, intended to improve the chance of survival on each side (Figure 1). A better understanding of host-pathogen interactions can elucidate unknown mechanisms, and therefore future targets, improving therapies of infectious diseases.
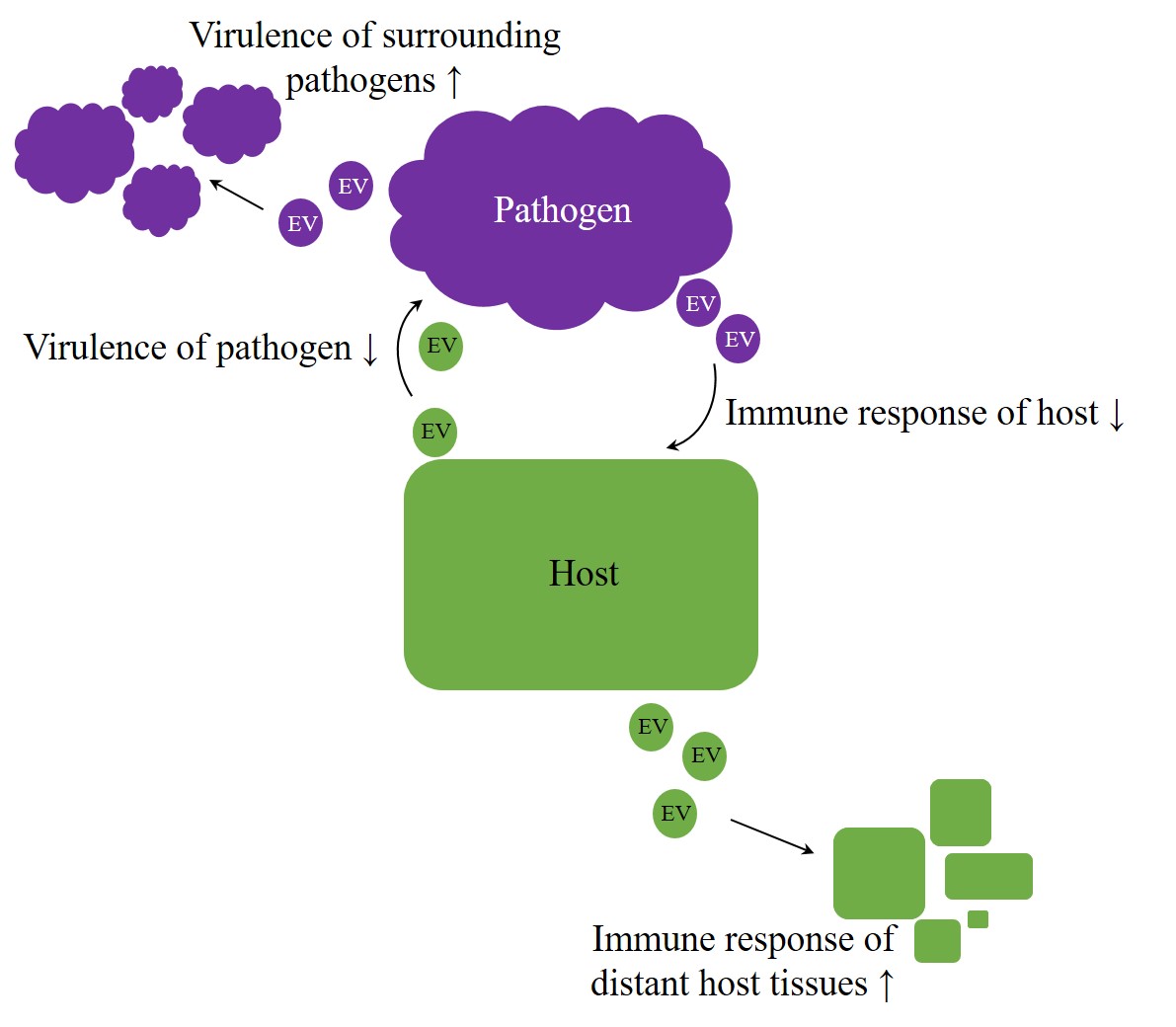
Figure 1. Arms race in host–pathogen interaction. Irrespective of kingdom boundaries, the genuine role of extracellular vesicles (EVs) appears to be bilateral. On the one hand, they were proven to have protective properties, but, on the other hand, they also appear to contribute to the achievement of inherent aims, like enhancing virulence on pathogens side or improving host´s immunity.
References
- Sager, R.; Palade, G.E. Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. Biophys. Biochem. Cytol. 1957, 3, 463–488, doi:10.1083/jcb.3.3.463.
- Sotelo, J.R.; Porter, K.R. An electron microscope study of the rat ovum. Biophys. Biochem. Cytol. 1959, 5, 327–342, doi:10.1083/jcb.5.2.327.
- De, S.N. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature 1959, 183, 1533–1534, doi:10.1038/1831533a0.
- Chatterjee, K.R.; Das Gupta, N.N.; De, M.L. Electron microscopic observations on the morphology of Mycobacterium leprae. Cell Res. 1959, 18, 521–527, doi:10.1016/0014-4827(59)90317-9.
- Jensen, W.A. The composition and ultrastructure of the nucellus in cotton. Ultrastruct. Res. 1965, 13, 112–128.
- Takeo, K.; Uesaka, I.; Uehira, K.; Nishiura, M. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. Bacteriol. 1973, 113, 1442–1448.
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. Cell Biol. 1983, 97, 329–339, doi:10.1083/jcb.97.2.329.
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. Cell Biol. 1985, 101, 942–948, doi:10.1083/jcb.101.3.942.
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). Biol. Chem. 1987, 262, 9412–9420.
- Thery, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15, doi:10.3410/B3-15.
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. Exp. Med. 1996, 183, 1161–1172, doi:10.1084/jem.183.3.1161.
- Prangishvili, D.; Holz, I.; Stieger, E.; Nickell, S.; Kristjansson, J.K.; Zillig, W. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. Bacteriol. 2000, 182, 2985–2988, doi:10.1128/jb.182.10.2985-2988.2000.
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Rev. Mol. Cell Biol. 2018, 19, 213–228, doi:10.1038/nrm.2017.125.
- Malloci, M.; Perdomo, L.; Veerasamy, M.; Andriantsitohaina, R.; Simard, G.; Martinez, M.C. Extracellular vesicles: Mechanisms in human health and disease. Redox Signal. 2019, 30, 813–856, doi:10.1089/ars.2017.7265.
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Cell Biol. 2007, 9, 654, doi:10.1038/ncb1596.
- Simons, M.; Raposo, G. Exosome—Vesicular carriers for intercellular communication. Opin. Cell Biol. 2009, 21, 575–581, doi:10.1016/j.ceb.2009.03.007.
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. BBA Gen. Subj. 2012, 1820, 940–948, doi:10.1016/j.bbagen.2012.03.017.
- Otsuka, K.; Yamamoto, Y.; Matsuoka, R.; Ochiya, T. Maintaining good miRNAs in the body keeps the doctor away: Perspectives on the relationship between food-derived natural products and microRNAs in relation to exosomes/extracellular vesicles. Nutr. Food Res. 2018, 62, 1700080, doi:10.1002/mnfr.201700080.
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297.
- Jiang, M.; Sang, X.; Hong, Z. Beyond nutrients: Food-derived microRNAs provide cross-kingdom regulation. Bioessays 2012, 34, 280–284, doi:10.1002/bies.201100181.
- Xie, W.Y.; Weng, A.; Melzig, M.F. MicroRNAs as new bioactive components in medicinal plants. Planta Med. 2016, 82, 1153–1162, doi:10.1055/s-0042-108450.
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. Biol. Chem. 2010, 285, 17442–17452, doi:10.1074/jbc.M110.107821.
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Cell 2010, 39, 133–144, doi:10.1016/j.molcel.2010.06.010.
- Koga, Y.; Yasunaga, M.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Matsumura, Y. Exosome can prevent RNase from degrading microRNA in feces. Gastrointest. Oncol. 2011, 2, 215–222, doi:10.3978/j.issn.2078-6891.2011.015.
- Keller, S.; Ridinger, J.; Rupp, A.K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. Transl. Med. 2011, 9, 86, doi:10.1186/1479-5876-9-86.
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Bihan, T.L.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Commun. 2014, 5, 5488.
- Rutter, B.D.; Innes, R.W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2016, 175, 728–741.
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. Nutr. 2016, 147, 3–10, doi:10.3945/jn.116.238949.
- Wolf, J.M.; Casadevall, A. Challenges posed by extracellular vesicles from eukaryotic microbes. Opin. Microbiol. 2014, 22, 73–78, doi:10.1016/j.mib.2014.09.012.
- Fuhrmann, G.; Neuer, A.L.; Herrmann, I.K. Extracellular vesicles—A promising avenue for the detection and treatment of infectious diseases? J. Pharm. Biopharm. 2017, 118, 56–61, doi:10.1016/j.ejpb.2017.04.005.
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Cell Biol. 2019, 21, 9–17, doi:10.1038/s41556-018-0250-9.
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Mol. Life Sci. 2011, 68, 2667–2688, doi:10.1007/s00018-011-0689-3.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. Cell Biol. 2013, 200, 373–383, doi:10.1083/jcb.201211138.
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. Extracell. Vesicles 2014, 3, 26913, doi:10.3402/jev.v3.26913.
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Extracell. Vesicles 2018, 7, 1535750, doi:10.1080/20013078.2018.1535750.
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials an ISEV position paper. Extracell. Vesicles 2015, 4, 30087.
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. Proteom. 2010, 73, 1907–1920, doi:10.1016/j.jprot.2010.06.006.
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Immunol. 2014, 5, 442, doi:10.3389/fimmu.2014.00442.
- Oksvold, M.P.; Neurauter, A.; Pedersen, K.W. Magnetic bead-based isolation of exosomes. In RNA Interference: Challenges and Therapeutic Opportunities; Sioud, M., Ed.; Springer: New York, NY, USA, 2015; pp. 465–481.
- Woo, J.; Sharma, S.; Gimzewski, J. The role of isolation methods on a nanoscale surface structure and its effect on the size of exosomes. Circ. Biomark. 2016, 5, 11, doi:10.5772/64148.
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Rev. Immunol. 2002, 2, 569–579, doi:10.1038/nri855.
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188, doi:10.1016/j.tcb.2016.11.003.
- Joshi, P.; Benussi, L.; Furlan, R.; Ghidoni, R.; Verderio, C. Extracellular vesicles in Alzheimer’s disease: Friends or foes? Focus on a beta-vesicle interaction. J. Mol. Sci. 2015, 16, 4800–4813, doi:10.3390/ijms16034800.
- Chen, Y.; Xiao, Y.; Lin, Z.; Xiao, X.; He, C.; Bihl, J.C.; Zhao, B.; Ma, X.; Chen, Y. The role of circulating platelets microparticles and platelet parameters in acute ischemic stroke patients. Stroke Cerebrovasc. Dis. 2015, 24, 2313–2320, doi:10.1016/j.jstrokecerebrovasdis.2015.06.018.
- Fais, S.; O’Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; da Silva, A.C.; Del Portillo, H.; El Andaloussi, S.; et al. Evidence-based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano 2016, 10, 3886–3899, doi:10.1021/acsnano.5b08015.
- McDonald, M.K.; Tian, Y.; Qureshi, R.A.; Gormley, M.; Ertel, A.; Gao, R.; Aradillas Lopez, E.; Alexander, G.M.; Sacan, A.; Fortina, P.; et al. Functional significance of macrophage-derived exosomes in inflammation and pain. Pain 2014, 155, 1527–1539, doi:10.1016/j.pain.2014.04.029.
- Katsiougiannis, S. Extracellular vesicles: Evolving Contributors in autoimmunity. Immunopathol. Dis. Therap. 2015, 6, 163–170, doi:10.1615/ForumImmunDisTher.2016016491.
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-derived exosomal miRNAs: A novel mechanism for obesity-related disease. Res. 2014, 77, 447–454.
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell derived exosomes. Med. 1998, 4, 594–600.
- Guo, W.; Gao, Y.; Li, N.; Shao, F.; Wang, C.; Wang, P.; Yang, Z.; Li, R.; He, J. Exosomes: New players in cancer (Review). Rep. 2017, 38, 665–675, doi:10.3892/or.2017.5714.
- Sullivan, R.; Maresh, G.; Zhang, X.; Salomon, C.; Hooper, J.; Margolin, D.; Li, L. The emerging roles of extracellular vesicles as communication vehicles within the tumor microenvironment and beyond. Endocrinol. (Lausanne) 2017, 8, 194, doi:10.3389/fendo.2017.00194.
- Zha, Q.B.; Yao, Y.F.; Ren, Z.J.; Li, X.J.; Tang, J.H. Extracellular vesicles: An overview of biogenesis, function, and role in breast cancer. Tumour Biol. 2017, 39, 1010428317691182, doi:10.1177/1010428317691182.
- Bastos, N.; Ruivo, C.F.; da Silva, S.; Melo, S.A. Exosomes in cancer: Use them or target them? Cell Dev. Biol. 2018, 78, 13–21, doi:10.1016/j.semcdb.2017.08.009.
- Kurywchak, P.; Tavormina, J.; Kalluri, R. The emerging roles of exosomes in the modulation of immune responses in cancer. Genome Med. 2018, 10, 23, doi:10.1186/s13073-018-0535-4.
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Med. 2013, 7, 769–778, doi:10.2217/bmm.13.63.
- Yanez-Mo, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. Extracell. Vesicles 2015, 4, 27066, doi:10.3402/jev.v4.27066.
- Wang, W.; Luo, J.; Wang, S. Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Healthc. Mater. 2018, 7, e1800484, doi:10.1002/adhm.201800484.
- Lai, P.; Weng, J.; Guo, L.; Chen, X.; Du, X. Novel insights into MSC-EVs therapy for immune diseases. Res. 2019, 7, 6.
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Cell Res. 2010, 4, 214–222, doi:10.1016/j.scr.2009.12.003.
- Lou, G.H.; Chen, Z.; Zheng, M.; Liu, Y.N. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Mol. Med. 2017, 49, e346, doi:10.1038/emm.2017.63.
- Adamiak, M.; Sahoo, S. Exosomes in myocardial repair: Advances and challenges in the development of next-generation therapeutics. Ther. 2018, 26, 1635–1643, doi:10.1016/j.ymthe.2018.04.024.
- Kordelas, L.; Rebmann, V.; Ludwig, A.K.; Radtke, S.; Ruesing, J.; Doeppner, T.R.; Epple, M.; Horn, P.A.; Beelen, D.W.; Giebel, B. MSC-derived exosomes: A novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 2014, 28, 970–973, doi:10.1038/leu.2014.41.
- Gimona, M.; Pachler, K.; Laner-Plamberger, S.; Schallmoser, K.; Rohde, E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. J. Mol. Sci. 2017, 18, 1190, doi:10.3390/ijms18061190.
- Palazzolo, S.; Bayda, S.; Hadla, M.; Caligiuri, I.; Corona, G.; Toffoli, G.; Rizzolio, F. The clinical translation of organic nanomaterials for cancer therapy: A focus on polymeric nanoparticles, micelles, liposomes and exosomes. Med. Chem. 2018, 25, 4224–4268, doi:10.2174/0929867324666170830113755.
- Piffoux, M.; Nicolas-Boluda, A.; Mulens-Arias, V.; Richard, S.; Rahmi, G.; Gazeau, F.; Wilhelm, C.; Silva, A.K.A. Extracellular vesicles for personalized medicine: The input of physically triggered production, loading and theranostic properties. Drug Deliv. Rev. 2019, 138, 247–258, doi:10.1016/j.addr.2018.12.009.
- Fuhrmann, G.; Chandrawati, R.; Parmar, P.A.; Keane, T.J.; Maynard, S.A.; Bertazzo, S.; Stevens, M.M. Engineering extracellular vesicles with the tools of enzyme prodrug therapy. Mater. 2018, 30, e1706616, doi:10.1002/adma.201706616.
- Kooijmans, S.A.A.; Fliervoet, L.A.L.; van der Meel, R.; Fens, M.; Heijnen, H.F.G.; van Bergen En Henegouwen, P.M.P.; Vader, P.; Schiffelers, R.M. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. Control. Release 2016, 224, 77–85, doi:10.1016/j.jconrel.2016.01.009.
- Murphy, D.E.; De Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Mol. Med. 2019, 51, 32, doi:10.1038/s12276-019-0223-5.
- Pinheiro, A.; Silva, A.M.; Teixeira, J.H.; Goncalves, R.M.; Almeida, M.I.; Barbosa, M.A.; Santos, S.G. Extracellular vesicles: Intelligent delivery strategies for therapeutic applications. Control. Release 2018, 289, 56–69, doi:10.1016/j.jconrel.2018.09.019.
- He, H.; Lu, Y.; Qi, J.; Zhu, Q.; Chen, Z.; Wu, W. Adapting liposomes for oral drug delivery. Acta Pharm. Sin. B 2019, 9, 36–48, doi:10.1016/j.apsb.2018.06.005.
- Wang, Q.; Zhuang, X.; Mu, J.; Deng, Z.B.; Jiang, H.; Zhang, L.; Xiang, X.; Wang, B.; Yan, J.; Miller, D.; et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Commun. 2013, 4, 1867, doi:10.1038/ncomms2886.
- Wang, X.; Zhang, M.; Flores, S.R.L.; Woloshun, R.R.; Yang, C.; Yin, L.; Xiang, P.; Xu, X.; Garrick, M.D.; Vidyasagar, S.; et al. Oral gavage of ginger nanoparticle-derived lipid vectors carrying Dmt1 siRNA blunts iron loading in murine hereditary hemochromatosis. Ther. 2019, 27, 493–506, doi:10.1016/j.ymthe.2019.01.003.
- Emam, S.E.; Ando, H.; Abu Lila, A.S.; Shimizu, T.; Ukawa, M.; Okuhira, K.; Ishima, Y.; Mahdy, M.A.; Ghazy, F.S.; Ishida, T. A novel strategy to increase the yield of exosomes (extracellular vesicles) for an expansion of basic research. Pharm. Bull. 2018, 41, 733–742, doi:10.1248/bpb.b17-00919.
- Palviainen, M.; Saari, H.; Karkkainen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R. Metabolic signature of extracellular vesicles depends on the cell culture conditions. Extracell. Vesicles 2019, 8, 1596669, doi:10.1080/20013078.2019.1596669.
- Adlerz, K.; Trempel, M.; Rowley, J.A.; Ahsan, T. Increasing yield of msc-evs in scalable xeno-free manufacturing. Cytotherapy 2019, 21, S58.
- Phan, J.; Kumar, P.; Hao, D.; Gao, K.; Farmer, D.; Wang, A. Engineering mesenchymal stem cells to improve their exosome efficacy and yield for cell-free therapy. Extracell. Vesicles 2018, 7, 1522236, doi:10.1080/20013078.2018.1522236.
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. Extracell. Vesicles 2018, 7, 1440132, doi:10.1080/20013078.2018.1440132.
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe 2018, 24, 637–652, e638, doi:10.1016/j.chom.2018.10.001.
- Regente, M.; Corti-Monzon, G.; Maldonado, A.M.; Pinedo, M.; Jorrin, J.; de la Canal, L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583, 3363–3366, doi:10.1016/j.febslet.2009.09.041.
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.B.; Wang, B.; Zhang, L.; et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Ther. 2013, 21, 1345–1357, doi:10.1038/mt.2013.64.
- Mu, J.Y.; Zhuang, X.Y.; Wang, Q.L.; Jiang, H.; Deng, Z.B.; Wang, B.M.; Zhang, L.F.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Nutr. Food Res. 2014, 58, 1561–1573, doi:10.1002/mnfr.201300729.
- Prado, N.; Alche Jde, D.; Casado-Vela, J.; Mas, S.; Villalba, M.; Rodriguez, R.; Batanero, E. Nanovesicles are secreted during pollen germination and pollen tube growth: A possible role in fertilization. Plant 2014, 7, 573–577, doi:10.1093/mp/sst153.
- Yang, J.; Hotz, T.; Broadnax, L.; Yarmarkovich, M.; Elbaz-Younes, I.; Hirschi, K.D. Anomalous uptake and circulatory characteristics of the plant-based small RNA MIR2911. Rep. 2016, 6, 26834, doi:10.1038/srep26834.
- Perez-Bermudez, P.; Blesa, J.; Soriano, J.M.; Marcilla, A. Extracellular vesicles in food: Experimental evidence of their secretion in grape fruits. J. Pharm. Sci. 2017, 98, 40–50, doi:10.1016/j.ejps.2016.09.022.
- Zhang, M.Z.; Viennois, E.; Xu, C.L.; Merlin, D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016, 4, e1134415, doi:10.1080/21688370.2015.1134415.
- Pocsfalvi, G.; Turiak, L.; Ambrosone, A.; Del Gaudio, P.; Puska, G.; Fiume, I.; Silvestre, T.; Vekey, K. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. Plant Physiol. 2018, 229, 111–121, doi:10.1016/j.jplph.2018.07.006.
- Woith, E.; Melzig, M.F. Extracellular vesicles from fresh and dried plants-simultaneous purification and visualization using gel electrophoresis. J. Mol. Sci. 2019, 20, 357, doi:10.3390/ijms20020357.
- Casadevall, A.; Nosanchuk, J.D.; Williamson, P.; Rodrigues, M.L. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009, 17, 158–162, doi:10.1016/j.tim.2008.12.005.
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Rev. Microbiol. 2015, 13, 620–630, doi:10.1038/nrmicro3480.
- Joffe, L.S.; Nimrichter, L.; Rodrigues, M.L.; Del Poeta, M. Potential roles of fungal extracellular vesicles during infection. mSphere 2016, 1, e00099–e00016, doi:10.1128/mSphere.00099-16.
- Regente, M.; Pinedo, M.; Clemente, H.S.; Balliau, T.; Jamet, E.; De la Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. Exp. Bot. 2017, 68, 5485–5495.
- Rutter, B.D.; Innes, R.W. Extracellular vesicles as key mediators of plant-microbe interactions. Opin. Plant Biol. 2018, 44, 16–22, doi:10.1016/j.pbi.2018.01.008.
- Walker, L.; Sood, P.; Lenardon, M.D.; Milne, G.; Olson, J.; Jensen, G.; Wolf, J.; Casadevall, A.; Adler-Moore, J.; Gow, N.A.R. The viscoelastic properties of the fungal cell wall allow traffic of ambisome as intact liposome vesicles. MBio 2018, 9, e02383, doi:10.1128/mBio.02383-17.
- Blackwood, R.A.; Smolen, J.E.; Transue, A.; Hessler, R.J.; Harsh, D.M.; Brower, R.C.; French, S. Phospholipase D activity facilitates Ca2+-induced aggregation and fusion of complex liposomes. J. Physiol. 1997, 272, C1279–C1285, doi:10.1152/ajpcell.1997.272.4.C1279.
- Zhang, M.Z.; Wang, X.Y.; Han, M.K.; Collins, J.F.; Merlin, D. Oral administration of ginger-derived nanolipids loaded with siRNA as a novel approach for efficient siRNA drug delivery to treat ulcerative colitis. Nanomedicine 2017, 12, 1927–1943, doi:10.2217/nnm-2017-0196.
- Wang, B.; Zhuang, X.; Deng, Z.B.; Jiang, H.; Mu, J.; Wang, Q.; Xiang, X.; Guo, H.; Zhang, L.; Dryden, G.; et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Ther. 2014, 22, 522–534, doi:10.1038/mt.2013.190.
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129, doi:10.1126/science.aar4142.
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Lo Dico, A.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527, doi:10.18632/oncotarget.4004.
- Record, M. Exosome-like nanoparticles from food: Protective nanoshuttles for bioactive cargo. Ther. 2013, 21, 1294–1296, doi:10.1038/mt.2013.130.
- Zhuang, X.; Deng, Z.B.; Mu, J.; Zhang, L.; Yan, J.; Miller, D.; Feng, W.; McClain, C.J.; Zhang, H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. Extracell. Vesicles 2015, 4, 28713, doi:10.3402/jev.v4.28713.
- Zhuang, X.Y.; Teng, Y.; Samykutty, A.; Mu, J.Y.; Deng, Z.B.; Zhang, L.F.; Cao, P.X.; Rong, Y.; Yan, J.; Miller, D.; et al. Grapefruit-derived nanovectors delivering therapeutic miR17 through an intranasal route inhibit brain tumor progression. Ther. 2016, 24, 96–105, doi:10.1038/mt.2015.188.
- Teng, Y.; Mu, J.; Hu, X.; Samykutty, A.; Zhuang, X.; Deng, Z.; Zhang, L.; Cao, P.; Yan, J.; Miller, D.; et al. Grapefruit-derived nanovectors deliver miR-18a for treatment of liver metastasis of colon cancer by induction of M1 macrophages. Oncotarget 2016, 7, 25683–25697, doi:10.18632/oncotarget.8361.
- Tang, Z.; Jun, Y.; Lv, Y.; Li, Y.; Zhang, Z.; Tao, M.; Chen, X.; He, J.; Zhang, L.; Wang, Q.-L. Aptamer-conjugated and doxorubicin-loaded grapefruit-derived nanovectors for targeted therapy against HER2+ breast cancer. Drug Target. 2019, 1–22, doi:10.1080/1061186X.2019.1624970.
- Zhang, M.Z.; Xiao, B.; Wang, H.; Han, M.K.; Zhang, Z.; Viennois, E.; Xu, C.L.; Merlin, D. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Ther. 2016, 24, 1783–1796, doi:10.1038/mt.2016.159.
- Lee, J.H.; Kim, Y.G.; Choi, P.; Ham, J.; Park, J.G.; Lee, J. Antibiofilm and antivirulence activities of 6-gingerol and 6-shogaol against candida albicans due to hyphal inhibition. Cell. Infect. Microbiol. 2018, 8, 299, doi:10.3389/fcimb.2018.00299.
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Plants 2016, 2, 16151, doi:10.1038/nplants.2016.151.
- Takeo, K.; Uesaka, I.; Uehira, K.; Nishiura, M. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. Bacteriol. 1973, 113, 1449–1454.
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Cell 2007, 6, 48–59, doi:10.1128/EC.00318-06.
- Oliveira, D.L.; Rizzo, J.; Joffe, L.S.; Godinho, R.M.C.; Rodrigues, M.L. Where do they come from and where do they go: Candidates for regulating extracellular vesicle formation in fungi. J. Mol. Sci. 2013, 14, 9581–9603, doi:10.3390/ijms14059581.
- Rodrigues, M.L.; Travassos, L.R.; Miranda, K.R.; Franzen, A.J.; Rozental, S.; de Souza, W.; Alviano, C.S.; Barreto-Bergter, E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Immun. 2000, 68, 7049–7060, doi:10.1128/iai.68.12.7049-7060.2000.
- Rodrigues, M.L.; Franzen, A.J.; Nimrichter, L.; Miranda, K. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Opin. Microbiol. 2013, 16, 414–420, doi:10.1016/j.mib.2013.04.002.
- Rodrigues, M.L.; Casadevall, A. A two-way road: Novel roles for fungal extracellular vesicles. Microbiol. 2018, 110, 11–15, doi:10.1111/mmi.14095.
- De Toledo Martins, S.; Szwarc, P.; Goldenberg, S.; Alves, L.R. Extracellular vesicles in fungi: Composition and functions. In Current Topics in Microbiology and Immunology; Ahmed, R., Akira, S., Aktories, K., Casadevall, A., Compans, R.W., Galan, J.E., Garcia-Sastre, A., Malissen, B., Rappuoli, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–15, doi:10.1007/82_2018_141.
- Peres da Silva, R.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; Cesar, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular vesicle-mediated export of fungal RNA. Rep. 2015, 5, 7763, doi:10.1038/srep07763.
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; das Neves, R.F.; Souto-Padron, T.; de Souza, W.; Cayota, A. Gene expression changes induced by Trypanosoma cruzi shed microvesicles in mammalian host cells: Relevance of tRNA-derived halves. Res. Int. 2014, 2014, 305239, doi:10.1155/2014/305239.
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43, doi:10.15252/embr.201439363.
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular transport in Histoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Microbiol. 2008, 10, 1695–1710, doi:10.1111/j.1462-5822.2008.01160.x.
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.; Miranda, K.; Silva, L.S.; Freymuller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-Galactosyl epitopes. Cell 2011, 10, 343–351, doi:10.1128/EC.00227-10.
- Vargas, G.; Rocha, J.D.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.; Medeiros, L.C.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Microbiol. 2015, 17, 389–407, doi:10.1111/cmi.12374.
- Yoneda, A.; Doering, T.L. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Biol. Cell 2006, 17, 5131–5140, doi:10.1091/mbc.E06-08-0701.
- Panepinto, J.; Komperda, K.; Frases, S.; Park, Y.D.; Djordjevic, J.T.; Casadevall, A.; Williamson, P.R. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Microbiol. 2009, 71, 1165–1176, doi:10.1111/j.1365-2958.2008.06588.x.
- Wolf, J.M.; Espadas, J.; Luque-Garcia, J.; Reynolds, T.; Casadevall, A. Lipid biosynthetic genes affect candida albicans extracellular vesicle morphology, cargo, and immunostimulatory properties. Cell 2015, 14, 745–754, doi:10.1128/EC.00054-15.
- Huang, S.H.; Wu, C.H.; Chang, Y.C.; Kwon-Chung, K.J.; Brown, R.J.; Jong, A. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS ONE 2012, 7, e48570, doi:10.1371/journal.pone.0048570.
- Gehrmann, U.; Qazi, K.R.; Johansson, C.; Hultenby, K.; Karlsson, M.; Lundeberg, L.; Gabrielsson, S.; Scheynius, A. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses—Novel mechanisms for host-microbe interactions in atopic eczema. PLoS ONE 2011, 6, e21480, doi:10.1371/journal.pone.0021480.
- Oliveira, D.L.; De-Lima, C.G.F.; Nosanchuk, J.D.; Casadevall, A.; Rodrigues, M.L.; Nimrichter, L. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Immun. 2010, 78, 1601–1609.
- Johansson, H.J.; Vallhov, H.; Holm, T.; Gehrmann, U.; Andersson, A.; Johansson, C.; Blom, H.; Carroni, M.; Lehtio, J.; Scheynius, A. Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Rep. 2018, 8, 9182, doi:10.1038/s41598-018-27451-9.
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.; Longo, L.V.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: Comparative analysis with other pathogenic fungi. Proteome Res. 2012, 11, 1676–1685, doi:10.1021/pr200872s.
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, e11113.
- Vallejo, M.C.; Nakayasu, E.S.; Longo, L.V.; Ganiko, L.; Lopes, F.G.; Matsuo, A.L.; Almeida, I.C.; Puccia, R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS ONE 2012, 7, e39463, doi:10.1371/journal.pone.0039463.
- Raj, S.; Nazemidashtarjandi, S.; Kim, J.; Joffe, L.; Zhang, X.; Singh, A.; Mor, V.; Desmarini, D.; Djordjevic, J.; Raleigh, D.P.; et al. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans. Biophys. Acta Biomembr. 2017, 1859, 2224–2233, doi:10.1016/j.bbamem.2017.08.017.
- Levery, S.B.; Momany, M.; Lindsey, R.; Toledo, M.S.; Shayman, J.A.; Fuller, M.; Brooks, K.; Doong, R.L.; Straus, A.H.; Takahashi, H.K. Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc:ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 2002, 525, 59–64, doi:10.1016/s0014-5793(02)03067-3.
- Rittenour, W.R.; Chen, M.; Cahoon, E.B.; Harris, S.D. Control of glucosylceramide production and morphogenesis by the Bar1 ceramide synthase in Fusarium graminearum. PLoS ONE 2011, 6, e19385, doi:10.1371/journal.pone.0019385.
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Rev. Microbiol. 2010, 64, 163–184, doi:10.1146/annurev.micro.091208.073413.
- Goes, A.; Fuhrmann, G. Biogenic and biomimetic carriers as versatile transporters to treat infections. Acs Infect Dis. 2018, 4, 881–892, doi:10.1021/acsinfecdis.8b00030.
- McBroom, A.J.; Kuehn, M.J. Outer membrane vesicles. EcoSal Plus 2005, 1, doi:10.1128/ecosal.2.2.4.
- Renelli, M.; Matias, V.; Lo, R.Y.; Beveridge, T.J. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004, 150, 2161–2169, doi:10.1099/mic.0.26841-0.
- Schulz, E.; Goes, A.; Garcia, R.; Panter, F.; Koch, M.; Muller, R.; Fuhrmann, K.; Fuhrmann, G. Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. Control. Release 2018, 290, 46–55, doi:10.1016/j.jconrel.2018.09.030.
- Arigita, C.; Jiskoot, W.; Westdijk, J.; Van Ingen, C.; Hennink, W.E.; Crommelin, D.J.; Kersten, G.F. Stability of mono- and trivalent meningococcal outer membrane vesicle vaccines. Vaccine 2004, 22, 629–642, doi:10.1016/j.vaccine.2003.08.027.
- Dauros Singorenko, P.; Chang, V.; Whitcombe, A.; Simonov, D.; Hong, J.; Phillips, A.; Swift, S.; Blenkiron, C. Isolation of membrane vesicles from prokaryotes: A technical and biological comparison reveals heterogeneity. Extracell. Vesicles 2017, 6, 1324731, doi:10.1080/20013078.2017.1324731.
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Carcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Commun. 2016, 7, 11220, doi:10.1038/ncomms11220.
- Schwechheimer, C.; Sullivan, C.J.; Kuehn, M.J. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 2013, 52, 3031–3040, doi:10.1021/bi400164t.
- McBroom, A.J.; Johnson, A.P.; Vemulapalli, S.; Kuehn, M.J. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. Bacteriol. 2006, 188, 5385–5392, doi:10.1128/Jb.00498-06.
- McBroom, A.J.; Kuehn, M.J. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Microbiol. 2006, 63, 545–558.
- Kulkarni, H.M.; Jagannadham, M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 2014, 160, 2109–2121, doi:10.1099/mic.0.079400-0.
- Elhenawy, W.; Bording-Jorgensen, M.; Valguarnera, E.; Haurat, M.F.; Wine, E.; Feldman, M.F. LPS remodeling triggers formation of outer membrane vesicles in salmonella. MBio 2016, 7, e00940-16, doi:10.1128/mBio.00940-16.
- Roier, S.; Leitner, D.R.; Iwashkiw, J.; Schild-Prufert, K.; Feldman, M.F.; Krohne, G.; Reidl, J.; Schild, S. Intranasal immunization with nontypeable Haemophilus influenzae outer membrane vesicles induces cross-protective immunity in mice. PLoS ONE 2012, 7, e42664, doi:10.1371/journal.pone.0042664.
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. Bacteriol. 1995, 177, 3998–4008, doi:10.1128/jb.177.14.3998-4008.1995.
- Bomberger, J.M.; MacEachran, D.P.; Coutermarsh, B.A.; Ye, S.; O´Toole, G.A.; Stanton, B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009, 5, e1000382.
- Kadurugamuwa, J.L.; Beveridge, T.J. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: Conceptually new antibiotics. Bacteriol. 1996, 178, 2767–2774, doi:10.1128/jb.178.10.2767-2774.1996.
- Schooling, S.R.; Beveridge, T.J. Membrane vesicles: An overlooked component of the matrices of biofilms. Bacteriol. 2006, 188, 5945–5957, doi:10.1128/JB.00257-06.
- Manning, A.J.; Kuehn, M.J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011, 11, 258, doi:10.1186/1471-2180-11-258.
- Ciofu, O.; Beveridge, T.J.; Kadurugamuwa, J.; Walther-Rasmussen, J.; Hoiby, N. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. Antimicrob. Chemother. 2000, 45, 9–13, doi:10.1093/jac/45.1.9.
- Chattopadhyay, M.K.; Jaganandham, M.V. Vesicles-mediated resistance to antibiotics in bacteria. Microbiol. 2015, 6, 758.
- Orench-Rivera, N.; Kuehn, M.J. Environmentally controlled bacterial vesicle-mediated export. Microbiol. 2016, 18, 1525–1536, doi:10.1111/cmi.12676.
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Microbiol. 2000, 37, 239–253, doi:10.1046/j.1365-2958.2000.01983.x.
- Jan, A.T. Outer membrane vesicles (OMVs) of gram-negative bacteria: A perspective update. Microbiol. 2017, 8, 1053, doi:10.3389/fmicb.2017.01053.
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005, 19, 2645–2655, doi:10.1101/gad.1299905.
- Wang, Y. The function of OmpA in Escherichia coli. Biophys. Res. Commun. 2002, 292, 396–401, doi:10.1006/bbrc.2002.6657.
- Bergman, M.A.; Cummings, L.A.; Barrett, S.L.; Smith, K.D.; Lara, J.C.; Aderem, A.; Cookson, B.T. CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Immun. 2005, 73, 1350–1356, doi:10.1128/IAI.73.3.1350-1356.2005.
- Menina, S.; Labouta, H.I.; Geyer, R.; Krause, T.; Gordon, S.; Dersch, P.; Lehr, C.M. Invasin-functionalized liposome nanocarriers improve the intracellular delivery of anti-infective drugs. RSC Adv. 2016, 6, 41622–41629, doi:10.1039/c6ra02988d.
- Ellen, A.F.; Albers, S.V.; Huibers, W.; Pitcher, A.; Hobel, C.F.; Schwarz, H.; Folea, M.; Schouten, S.; Boekema, E.J.; Poolman, B.; et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009, 13, 67–79, doi:10.1007/s00792-008-0199-x.
- Rachel, R.; Wyschkony, I.; Riehl, S.; Huber, H. The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 2002, 1, 9–18.
- Gaudin, M.; Gauliard, E.; Schouten, S.; Houel-Renault, L.; Lenormand, P.; Marguet, E.; Forterre, P. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Microbiol. Rep. 2013, 5, 109–116, doi:10.1111/j.1758-2229.2012.00348.x.
- Erdmann, S.; Tschitschko, B.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Microbiol. 2017, 2, 1446–1455, doi:10.1038/s41564-017-0009-2.
- Ellen, A.F.; Rohulya, O.V.; Fusetti, F.; Wagner, M.; Albers, S.V.; Driessen, A.J. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. Bacteriol. 2011, 193, 4380–4387, doi:10.1128/JB.05028-11.
- Caspi, Y.; Dekker, C. Dividing the Archaeal Way: The Ancient Cdv Cell-Division Machinery. Microbiol. 2018, 9, 174, doi:10.3389/fmicb.2018.00174.
- Soler, N.; Marguet, E.; Verbavatz, J.M.; Forterre, P. Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Microbiol. 2008, 159, 390–399, doi:10.1016/j.resmic.2008.04.015.
- Gaudin, M.; Krupovic, M.; Marguet, E.; Gauliard, E.; Cvirkaite-Krupovic, V.; Le Cam, E.; Oberto, J.; Forterre, P. Extracellular membrane vesicles harbouring viral genomes. Microbiol. 2014, 16, 1167–1175, doi:10.1111/1462-2920.12235.
- Tong, M.; Chamley, L.W. Placental extracellular vesicles and feto-maternal communication. Cold Spring Harb. Perspect. Med. 2015, 5, a023028, doi:10.1101/cshperspect.a023028.
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. Transl. Med. 2011, 9, 9, doi:10.1186/1479-5876-9-9.
- Zonneveld, M.I.; Brisson, A.R.; van Herwijnen, M.J.C.; Tan, S.; van de Lest, C.H.A.; Redegeld, F.A.; Garssen, J.; Wauben, M.H.M.; Hoen, E.N.M.N.-t. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. Extracell. Vesicles 2014, 3, 24215, doi:10.3402/jev.v3.24215.
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Nosanchuk, J.D.; Casadevall, A. Vesicular Trans-Cell Wall Transport in Fungi: A Mechanism for the Delivery of Virulence-Associated Macromolecules? Lipid Insights 2008, 2, 27–40, doi:10.4137/lpi.s1000.
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’Donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Commun. 2018, 9, 1556, doi:10.1038/s41467-018-03991-6.
- Ellis, T.N.; Kuehn, M.J. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Mol. Biol. Rev. 2010, 74, 81–94, doi:10.1128/MMBR.00031-09.
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. Nutr. 2014, 144, 1495–1500, doi:10.3945/jn.114.196436.
- Wolf, T.; Baier, S.R.; Zempleni, J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma CaCo-2 cells and rat small intestinal IEC-6 cells. Nutr. 2015, 145, 2201–2206, doi:10.3945/jn.115.218586.
- Bernal, D.; Trelis, M.; Montaner, S.; Cantalapiedra, F.; Galiano, A.; Hackenberg, M.; Marcilla, A. Surface analysis of Dicrocoelium dendriticum. The molecular characterization of exosomes reveals the presence of miRNAs. Proteom. 2014, 105, 232–241, doi:10.1016/j.jprot.2014.02.012.
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Environ. Microbiol. 2000, 66, 4414–4420, doi:10.1128/aem.66.10.4414-4420.2000.
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123, doi:10.1126/science.1239705.
- Weiberg, A.; Jin, H. Small RNAs—The secret agents in the plant-pathogen interactions. Opin. Plant Biol. 2015, 26, 87–94, doi:10.1016/j.pbi.2015.05.033.
- Wang, M.; Weiberg, A.; Dellota, E., Jr.; Yamane, D.; Jin, H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428, doi:10.1080/15476286.2017.1291112.
- Macdonald, I.A.; Kuehn, M.J. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. Bacteriol. 2013, 195, 2971–2981, doi:10.1128/JB.02267-12.
- Rivera, J.; Cordero, R.J.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Natl. Acad. Sci. USA 2010, 107, 19002–19007, doi:10.1073/pnas.1008843107.
- Kaparakis, M.; Turnbull, L.; Carneiro, L.; Firth, S.; Coleman, H.A.; Parkington, H.C.; Le Bourhis, L.; Karrar, A.; Viala, J.; Mak, J.; et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Microbiol. 2010, 12, 372–385, doi:10.1111/j.1462-5822.2009.01404.x.
- Deo, P.; Chow, S.H.; Hay, I.D.; Kleifeld, O.; Costin, A.; Elgass, K.D.; Jiang, J.H.; Ramm, G.; Gabriel, K.; Dougan, G.; et al. Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog. 2018, 14, e1006945, doi:10.1371/journal.ppat.1006945.
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A.; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016, 12, e1005672, doi:10.1371/journal.ppat.1005672.
- Codemo, M.; Muschiol, S.; Iovino, F.; Nannapaneni, P.; Plant, L.; Wai, S.N.; Henriques-Normark, B. Immunomodulatory effects of pneumococcal extracellular vesicles on cellular and humoral host defenses. MBio 2018, 9, e00559-18, doi:10.1128/mBio.00559-18.
- Poon, I.K.H.; Gregory, C.D.; Kaparakis-Liaskos, M. Editorial: The immunomodulatory properties of extracellular vesicles from pathogens, immune cells, and non-immune cells. Immunol. 2018, 9, 3024, doi:10.3389/fimmu.2018.03024.
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126, doi:10.1038/cr.2011.158.
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006, doi:10.1038/cr.2008.282.
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Natl. Acad. Sci. USA 2008, 105, 10513–10518, doi:10.1073/pnas.0804549105.
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7.
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. Dairy Sci. 2012, 95, 4831–4841, doi:10.3168/jds.2012-5489.
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. J. Environ. Res. Public Health 2015, 12, 13981–14020, doi:10.3390/ijerph121113981.
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Biol. 2005, 15, 1501–1507, doi:10.1016/j.cub.2005.07.029.
- Xie, W.; Melzig, M.F. The stability of medicinal plant microRNAs in the herb preparation process. Molecules 2018, 23, 919, doi:10.3390/molecules23040919.
- Howard, K.M.; Jati Kusuma, R.; Baier, S.R.; Friemel, T.; Markham, L.; Vanamala, J.; Zempleni, J. Loss of miRNAs during processing and storage of cow’s (Bos taurus) milk. Agric. Food Chem. 2015, 63, 588–592, doi:10.1021/jf505526w.
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108, doi:10.1186/1471-2407-11-108.
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. Extracell. Vesicles 2014, 3, 24641, doi:10.3402/jev.v3.24641.
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61, doi:10.1016/j.canlet.2015.10.020.
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. Immunol. 2007, 179, 1969–1978, doi:10.4049/jimmunol.179.3.1969.
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137, doi:10.1038/cr.2010.80.
- Hata, T.; Murakami, K.; Nakatani, H.; Yamamoto, Y.; Matsuda, T.; Aoki, N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biophys. Res. Commun. 2010, 396, 528–533, doi:10.1016/j.bbrc.2010.04.135.
- Chen, T.; Xie, M.Y.; Sun, J.J.; Ye, R.S.; Cheng, X.; Sun, R.P.; Wei, L.M.; Li, M.; Lin, D.L.; Jiang, Q.Y.; et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Rep. 2016, 6, 33862, doi:10.1038/srep33862.
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854, doi:10.1038/27579.
- Newmark, P.A.; Reddien, P.W.; Cebria, F.; Alvarado, A.S. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Natl. Acad. Sci. USA 2003, 100, 11861–11865.
- Issa, Z.; Grant, W.N.; Stasiuk, S.; Shoemaker, C.B. Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis. J. Parasitol. 2005, 35, 935–940, doi:10.1016/j.ijpara.2005.06.001.
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. Insect Physiol. 2011, 57, 231–245, doi:10.1016/j.jinsphys.2010.11.006.
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Plant Sci. 2015, 6, 766, doi:10.3389/fpls.2015.00766.
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Plants 2016, 2, 16153, doi:10.1038/nplants.2016.153.
- Castillo-Gonzalez, C.; Zhang, X.R. The Trojan horse of the plant kingdom. Cell Host Microbe 2018, 24, 1–3, doi:10.1016/j.chom.2018.06.015.
- Jain, S.; Pillai, J. Bacterial membrane vesicles as novel nanosystems for drug delivery. J. Nanomed. 2017, 12, 6329–6341, doi:10.2147/Ijn.S137368.
- Kaparakis-Liaskos, M.; Ferrero, R.L. Immune modulation by bacterial outer membrane vesicles. Rev. Immunol. 2015, 15, 375–387, doi:10.1038/nri3837.
- Kim, O.Y.; Hong, B.S.; Park, K.S.; Yoon, Y.J.; Choi, S.J.; Lee, W.H.; Roh, T.Y.; Lotvall, J.; Kim, Y.K.; Gho, Y.S. Immunization with Escherichia coli outer membrane vesicles protects bacteria-induced lethality via Th1 and Th17 cell responses. Immunol. 2013, 190, 4092–4102, doi:10.4049/jimmunol.1200742.
- Silverman, J.M.; Clos, J.; Horakova, E.; Wang, A.Y.; Wiesgigl, M.; Kelly, I.; Lynn, M.A.; McMaster, W.R.; Foster, L.J.; Levings, M.K.; et al. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. Immunol. 2010, 185, 5011–5022, doi:10.4049/jimmunol.1000541.
- Khowawisetsut, L. Extracellular vesicles in malaria infection. Siriraj Med. J. 2019, 71, 89–94.
- Twu, O.; de Miguel, N.; Lustig, G.; Stevens, G.C.; Vashisht, A.A.; Wohlschlegel, J.A.; Johnson, P.J. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate host: Parasite interactions. PLoS Pathog. 2013, 9, e1003482, doi:10.1371/journal.ppat.1003482.
- Marcilla, A.; Martin-Jaular, L.; Trelis, M.; De Menezes-Neto, A.; Osuna, A.; Bernal, D.; Fernandez-Becerra, C.; Almeida, I.C.; Del Portillo, H.A. Extracellular vesicles in parasitic diseases. Extracell. Vesicles 2014, 3, 25040, doi:10.3402/jev.v3.25040.
- Kuipers, M.E.; Hokke, C.H.; Smits, H.H.; Hoene, E.N.M.N. Pathogen-derived extracellular vesicle-associated molecules that affect the host immune system: An overview. Microbiol. 2018, 9, 2182, doi:10.3389/fmicb.2018.02182.
- Mekonnen, G.G.; Pearson, M.; Loukas, A.; Sotillo, J. Extracellular vesicles from parasitic helminths and their potential utility as vaccines. Expert Rev. Vaccines 2018, 17, 197–205, doi:10.1080/14760584.2018.1431125.
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. Control. Release 2015, 205, 35–44, doi:10.1016/j.jconrel.2014.11.029.
- Stremersch, S.; De Smedt, S.C.; Raemdonck, K. Therapeutic and diagnostic applications of extracellular vesicles. Control. Release 2016, 244, 167–183, doi:10.1016/j.jconrel.2016.07.054.
