NCAM2 governs neuronal morphogenesis and axodendritic architecture, and controls important neuron-specific processes such as neuronal differentiation, synaptogenesis and memory formation. In the adult brain, NCAM2 is highly expressed in dendritic spines, and it regulates synaptic plasticity and learning processes. NCAM2’s functions are related to its ability to adapt to the external inputs of the cell and to modify the cytoskeleton accordingly. Different studies show that NCAM2 interacts with proteins involved in cytoskeleton stability and proteins that regulate calcium influx, which could also modify the cytoskeleton.
- NCAM2
- cytoskeleton
- Actin
- microtubule
- MAP2
- CaMKII
- Autism Spectrum Disorder
- Alzheimer’s disease
1. Introduction
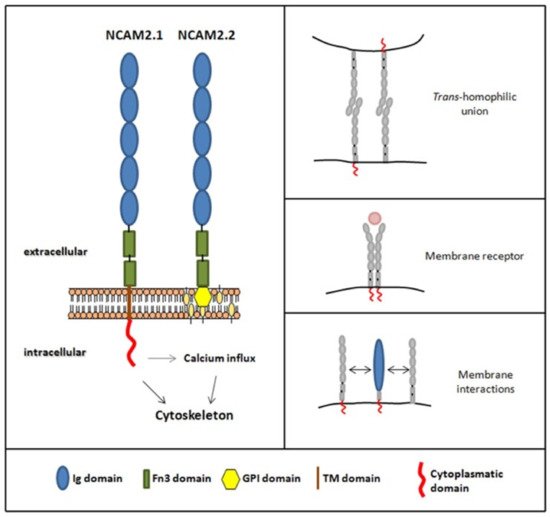
2. NCAM2 Expression and Interactors
3. NCAM2 in Neuronal Cell Fate Determination and Differentiation
| Extracellular Region | |
|---|---|
| Interaction | References |
| Fibroblast Growth Factor Receptor (FGFR) | [20] |
| Epidermal Growth Factor Receptor (EGFR) | [19] |
| Nogo | [9] |
| Granulin | [9] |
| Prion protein (Prp) | [21][22] |
| Beta-site APP cleaving enzyme 1 (BACE1) | [15] |
| Post-translational modification | |
| N-glycosylation | [18] |
| Proteolytic cleavage | [15][16] |
| Intracellular region | |
| Interaction | References |
| Proto-oncogene tyrosine-protein kinase Src | [23] |
| Calcium/calmodulin-dependent protein kinase type II (CaMKII) | [8][23] |
| Microtubule-associated protein 2 (MAP2) | [8] |
| Actin | [9] |
| Tubulin | [8] |
| 14-3-3 family proteins | [8][21][22] |
| Microtubule-associated protein 1B (MAP1B) | [9] |
| F-actin-capping complex (CAPZ) | [9] |
| Heat shock cognate 71 protein (HSC70) | [9] |
| Postranslational modification | |
| Palmitoylation | [9][14] |
| Phosphorylation | [13] |
3. NCAM2 in Neuronal Cell Fate Determination and Differentiation
During neuronal development, the differentiation of a neuron from a postmitotic cell encompasses different processes [84][24]. Imbalances in the homeostasis of the cytoskeleton dynamics of the postmitotic cell give rise to neuronal polarization, which results in the formation of dendrites and an axonal terminal [85,86,87][25][26][27]. This polarization process is intrinsic to the differentiating neuron but modulated through external signals. Different CAMs are involved in neuronal polarization, act as transducers with the outside, and are key in the overall neuronal development and differentiation processes both in vivo and in vitro [14,88][28][29]. Studies with NCAM2 revealed that this protein is essential in the process of neuronal differentiation [69,83][8][23]. The distribution of NCAM2 inside the neuron and its interactions with proteins that regulate the cytoskeleton dynamics are key to NCAM2 participating in the establishment of both the dendritic tree and the axonal process.
3.1. NCAM2 Role in Neuronal Migration and Corticogenesis
Brain formation involves a myriad of mechanisms, including temporal and spatial regulations during neuronal development that only occur in vivo [14][28]. The study of the mechanisms that regulate the polarization and development of neurons in vitro made it possible to identify proteins that have an implication at the physiological level [89][30]. Not only that, but the complexity of in vivo models has demonstrated the relevance of the extracellular matrix and cell adhesion molecules in the overall development of the brain. In this sense, adhesion proteins participate in different processes of neuronal development, such as neuronal migration, positioning, morphogenesis, development of dendritic and axonal compartments, and synaptogenesis [58,88,90,91][31][29][32][33]. In vivo, NCAM2 expression progressively increases during neuronal development; NCAM2 is involved in neuronal migration, cell positioning during corticogenesis, neuronal differentiation and synaptogenesis in the olfactory bulb, neocortex and hippocampus. In these regions, NCAM2 participates in the development of dendrites and axons, the establishment of connections, and the regulation of synaptic plasticity [66[5][7][34],68,92], see Figure 2.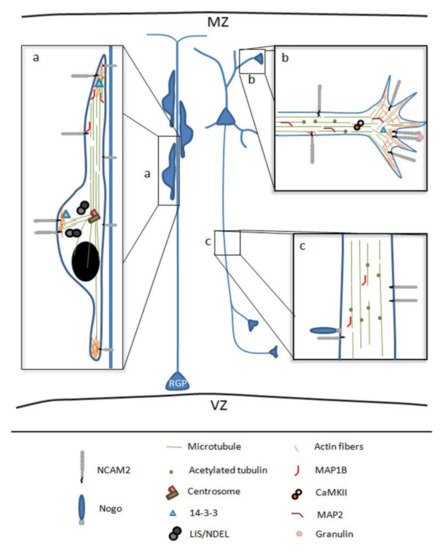
3.2. Neuronal Differentiation
3.2.1. NCAM2 in Dendritic Tree Development
3.2.2. NCAM2 in Axon Formation and Development
Different mechanisms are involved in the branching and elongation of the axonal terminal, some of which required a certain level of cytoskeleton stability [122][35]. In this sense, excessive instability of the microtubule cytoskeleton results in a higher number of branching points [123][36]. NCAM2 deficiency increases the number of branches and reduces the maximum length of the main axon, without altering the total length of the axonal shaft [69][8]. Moreover, 20% of NCAM2-depleted neurons exhibit two or more axons. At the cytoskeleton level, NCAM2 deficiency reduces the signal of acetylated tubulin in axons. Tubulin acetylation is a posttranslational modification that occurs only when microtubules are polymerized and thus is an indirect measure of microtubule cytoskeleton stability [124][37]. Regarding axon elongation, NCAM2 interacts with MAP1B, which binds to the microtubules and regulates their dynamics. The regulation of MAP1B binding to microtubules is crucial during the axon branching process and changes according to the degree of phosphorylation [125][38]. In addition, NCAM2 interactions with the light- and medium subunits of neurofilaments have been identified. During development and neuritogenesis, these two subunits form heterodimers and participate in the cytoskeleton dynamics required for axonal growth and transport [126][39]. Through direct interaction with these neurofilament subunits, with MAP1B, or through signaling pathways that modulate said stability, NCAM2 has a role in stabilizing microtubule cytoskeleton to ensure a proper axon development in terms of length and branching points.3.2.3. NCAM2 in Synaptogenesis and Synaptic Plasticity
During brain development, neuronal polarization and synaptogenesis occur in parallel. The process of stabilization of synaptic contacts is dependent on cytoskeleton rearrangement and neuronal activity. Synaptogenesis is highly regulated by cell adhesion molecules, which control the recognition of membranes, allow the formation of synaptic structures and modulate the neuronal maturation through calcium signaling [132,133,134,135][40][41][42][43]. In vivo, NCAM2 is detected in presynaptic and postsynaptic compartments and presents trans-homophilic binding between both compartments [68][7]—see Figure 3.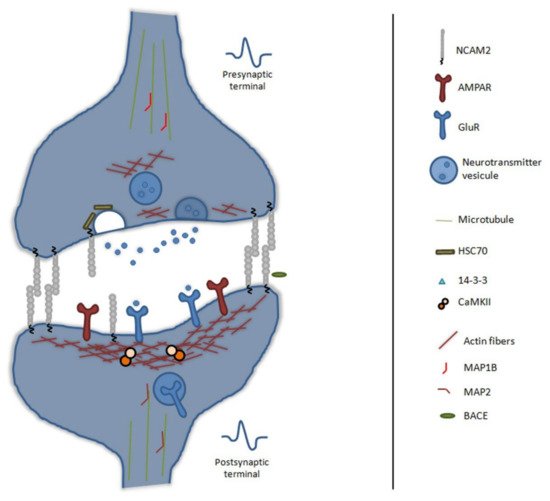
3.3. NCAM2 in Calcium Signaling and Homeostasis
4. NCAM2 in Neuronal Diseases
NCAM2 is expressed during brain development as well as in adulthood and its functions are crucial for the proper cognitive process. Genetic and molecular studies show that changes in NCAM2 expression could be the cause of different pathologies both during brain development and in the adult stages—see Table 32.| Neurodevelopmental Disorder | |||
|---|---|---|---|
| Disorder | Type of Study | Implications | References |
| Autism Spectrum Disorders | Genetic in humans | Genetic studies associate alterations and deletions in Ncam2 with ASD. | [47][48][49] |
| Down Syndrome | Genetic in humans | Increased expression in DS patients due to the location of Ncam2 in the 21 chromosome. | [6][50] |
| Other neurodevelopmental disorders | Genetic in humans | Deletions Ncam2 are found in patients with neurodevelpmental disorders. | [51] |
| Neurodegenerative diseases | |||
| Disorder | Implications | References | |
| Alzheimer’s Disease | Genetic in humans | Alterations in Ncam2 found in AD patients. | [52][53] |
| Experimental with human and mouse samples. | β-amyloid induces proteolysis of synaptic NCAM2. | [16] | |
| Frontotemporal dementia | Experimental with mouse tissue samples | NCAM2 proposed as a candidate receptor for GRN | [9] |
4.1. Neurodevelopment Diseases
4.1. Neurodevelopment Diseases
4.2. Neurodegenerative Diseases
5. Conclusions
In the development of the nervous system, NCAM2 regulates the molecular recognition that allows the formation of contacts between axonal and dendritic compartments leading to the formation of synapses and neural circuits. In particular, NCAM2 is present in both axonal and dendritic compartments and establishes homophilic junctions and intracellular interactions. These interactions regulate the modification of the cytoskeleton and affect the structure of the dendrite and axon. Different mechanisms are modulated by NCAM2 interactions but these can be divided in two groups: calcium signaling mechanisms and structuring of the cytoskeleton. NCAM2 is essential for the proper development of dendritic and axonal structures; its depletion causes aberrant neuron migration and differentiation and is detected in neurodevelopmental diseases. With these results, we can affirm that changes in NCAM2 alter the neuronal molecular identity, produces alterations in neuronal development and conduces to an aberrant neuronal network formation. Moreover, different CAMs modulate the neuronal differentiation, structure pruning and synaptic stabilization through the activation of gene expression. It is known that calcium influx and CaMKII modulate gene expression. More studies are necessary to identify the possible role of NCAM2 in gene expression (Figure 4).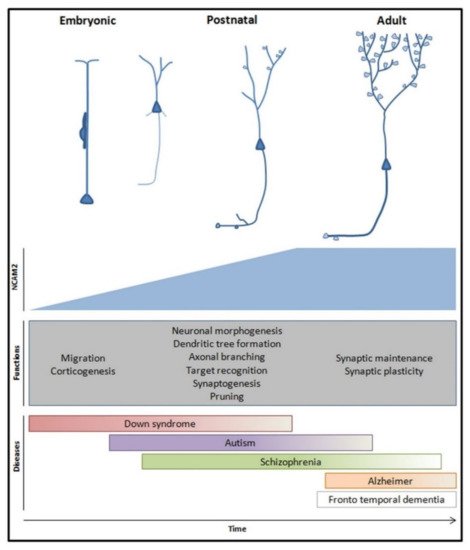
References
- Pebusque, M.J.; Coulier, F.; Birnbaum, D.; Pontarotti, P. Ancient large-scale genome duplications: Phylogenetic and linkage analyses shed light on chordate genome evolution. Mol. Biol. Evol. 1998, 15, 1145–1159.
- Makino, T.; McLysaght, A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc. Natl. Acad. Sci. USA 2010, 107, 9270–9274.
- Kiselyov, V.V.; Skladchikova, G.; Hinsby, A.M.; Jensen, P.H.; Kulahin, N.; Soroka, V.; Pedersen, N.; Tsetlin, V.; Poulsen, F.M.; Berezin, V.; et al. Structural Basis for a Direct Interaction between FGFR1 and NCAM and Evidence for a Regulatory Role of ATP. Structure 2003, 11, 691–701.
- Alenius, M.; Bohm, S. Identification of a Novel Neural Cell Adhesion Molecule-related Gene with a Potential Role in Selective Axonal Projection. J. Biol. Chem. 1997, 272, 26083–26086.
- Yoshihara, Y.; Kawasaki, M.; Tamada, A.; Fujita, H.; Hayashi, H.; Kagamiyama, H.; Mori, K. OCAM: A New Member of the Neural Cell Adhesion Molecule Family Related to Zone-to-Zone Projection of Olfactory and Vomeronasal Axons. J. Neurosci. 1997, 17, 5830–5842.
- Paoloni-Giacobino, A.; Chen, H.; Antonarakis, S.E. Cloning of a Novel Human Neural Cell Adhesion Molecule Gene (NCAM2) That Maps to Chromosome Region 21q21 and Is Potentially Involved in Down Syndrome. Genomics 1997, 43, 43–51.
- Ichinohe, N.; Yoshihara, Y.; Hashikawa, T.; Rockland, K.S. Developmental study of dendritic bundles in layer 1 of the rat granular retrosplenial cortex with special reference to a cell adhesion molecule, OCAM. Eur. J. Neurosci. 2003, 18, 1764–1774.
- Parcerisas, A.; Pujadas, L.; Ortega-Gascó, A.; Perelló-Amorós, B.; Viais, R.; Hino, K.; Figueiro-Silva, J.; La Torre, A.; Trullás, R.; Simó, S.; et al. NCAM2 Regulates Dendritic and Axonal Differentiation through the Cytoskeletal Proteins MAP2 and 14-3-3. Cereb. Cortex 2020, 30, 3781–3799.
- Parcerisas, A.; Ortega-Gascó, A.; Hernaiz-Llorens, M.; Odena, M.A.; Ulloa, F.; de Oliveira, E.; Bosch, M.; Pujadas, L.; Soriano, E. New Partners Identified by Mass Spectrometry Assay Reveal Functions of NCAM2 in Neural Cytoskeleton Organization. Int. J. Mol. Sci. 2021, 22, 7404.
- Maness, P.F.; Schachner, M. Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2006, 10, 19–26.
- Niethammer, P.; Delling, M.; Sytnyk, V.; Dityatev, A.; Fukami, K.; Schachner, M. Cosignaling of NCAM via lipid rafts and the FGF receptor is required for neuritogenesis. J. Cell Biol. 2002, 157, 521–532.
- Kamiguchi, H. The region-specific activities of lipid rafts during axon growth and guidance. J. Neurochem. 2006, 98, 330–335.
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villen, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A Tissue-Specific Atlas of Mouse Protein Phosphorylation and Expression. Cell 2010, 143, 1174–1189.
- Kulahin, N.; Walmod, P.S. The Neural Cell Adhesion Molecule NCAM2/OCAM/RNCAM, a Close Relative to NCAM. In Structure and Function of the Neural Cell Adhesion Molecule NCAM; Springer: New York, NY, USA, 2009; Volume 663, pp. 403–420.
- Kim, W.H.; Watanabe, H.; Lomoio, S.; Tesco, G. Spatiotemporal processing of neural cell adhesion molecules 1 and 2 by BACE1 in vivo. J. Biol. Chem. 2021, 296, 100372.
- Leshchyns’Ka, I.; Liew, H.T.; Shepherd, C.; Halliday, G.M.; Stevens, C.H.; Ke, Y.D.; Ittner, L.M.; Sytnyk, V. Aβ-dependent reduction of NCAM2-mediated synaptic adhesion contributes to synapse loss in Alzheimer’s disease. Nat. Commun. 2015, 6, 8836.
- Yu, R.K.; Yanagisawa, M. Glycobiology of neural stem cells. CNS Neurol. Disord. Drug Targets 2006, 5, 415–423.
- Kulahin, N.; Kristensen, O.; Rasmussen, K.K.; Olsen, L.; Rydberg, P.; Vestergaard, B.; Kastrup, J.S.; Berezin, V.; Bock, E.; Walmod, P.S.; et al. Structural Model and trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule. Structure 2011, 19, 203–211.
- Deleyrolle, L.; Sabourin, J.-C.; Rothhut, B.; Fujita, H.; Guichet, P.-O.; Teigell, M.; Ripoll, C.; Chauvet, N.; Perrin, F.; Mamaeva, D.; et al. OCAM Regulates Embryonic Spinal Cord Stem Cell Proliferation by Modulating ErbB2 Receptor. PLoS ONE 2015, 10, e0122337.
- Rasmussen, K.K.; Falkesgaard, M.H.; Winther, M.; Roed, N.K.; Quistgaard, C.L.; Teisen, M.N.; Edslev, S.M.; Petersen, D.L.; Aljubouri, A.; Christensen, C.; et al. NCAM2 Fibronectin type-III domains form a rigid structure that binds and activates the Fibroblast Growth Factor Receptor. Sci. Rep. 2018, 8, 8957.
- Collins, M.; Husi, H.; Yu, L.; Brandon, J.M.; Anderson, C.N.G.; Blackstock, W.P.; Choudhary, J.; Grant, S. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 2006, 97, 16–23.
- Kislinger, T.; Cox, B.; Kannan, A.; Chung, C.; Hu, P.; Ignatchenko, A.; Scott, M.S.; Gramolini, A.O.; Morris, Q.; Hallett, M.T.; et al. Global Survey of Organ and Organelle Protein Expression in Mouse: Combined Proteomic and Transcriptomic Profiling. Cell 2006, 125, 173–186.
- Sheng, L.; Leshchyns’Ka, I.; Sytnyk, V. Neural Cell Adhesion Molecule 2 Promotes the Formation of Filopodia and Neurite Branching by Inducing Submembrane Increases in Ca2+ Levels. J. Neurosci. 2015, 35, 1739–1752.
- Urbanska, M.; Blazejczyk, M.; Jaworski, J. Molecular Basis of Dendritic Arborization. Available online: https://pubmed.ncbi.nlm.nih.gov/18511961/ (accessed on 3 August 2021).
- Da Silva, J.S.; Dotti, C.G. Breaking the neuronal sphere: Regulation of the actin cytoskeleton in neuritogenesis. Nat. Rev. Neurosci. 2002, 3, 694–704.
- Cheng, P.-L.; Poo, M.-M. Early Events in Axon/Dendrite Polarization. Annu. Rev. Neurosci. 2012, 35, 181–201.
- Takano, T.; Xu, C.; Funahashi, Y.; Namba, T.; Kaibuchi, K. Neuronal polarization. Development 2015, 142, 2088–2093.
- Namba, T.; Funahashi, Y.; Nakamuta, S.; Xu, C.; Takano, T.; Kaibuchi, K. Extracellular and Intracellular Signaling for Neuronal Polarity. Physiol. Rev. 2015, 95, 995–1024.
- Kawauchi, T. Cell Adhesion and Its Endocytic Regulation in Cell Migration during Neural Development and Cancer Metastasis. Int. J. Mol. Sci. 2012, 13, 4564–4590.
- Farah, C.A.; Leclerc, N. HMWMAP2: New perspectives on a pathway to dendritic identity. Cell Motil. Cytoskelet. 2008, 65, 515–527.
- Sytnyk, V.; Leshchyns’Ka, I.; Schachner, M. Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends Neurosci. 2017, 40, 295–308.
- Dalva, M.B.; McClelland, A.C.; Kayser, M.S. Cell adhesion molecules: Signalling functions at the synapse. Nat. Rev. Neurosci. 2007, 8, 206–220.
- Seong, E.; Yuan, L.; Arikkath, J. Cadherins and catenins in dendrite and synapse morphogenesis. Cell Adhes. Migr. 2015, 9, 202–213.
- Ichinohe, N.; Knight, A.; Ogawa, M.; Ohshima, T.; Mikoshiba, K.; Yoshihara, Y.; Terashima, T.; Rockland, K. Unusual Patch-Matrix Organization in the Retrosplenial Cortex of the reeler Mouse and Shaking Rat Kawasaki. Cereb. Cortex 2008, 18, 1125–1138.
- Katherine, K.; Dent, W.E. Developing vertebrate CNS. Nat. Rev. Neurosci. 2014, 15, 7–18.
- Homma, N.; Takei, Y.; Tanaka, Y.; Nakata, T.; Terada, S.; Kikkawa, M.; Noda, Y.; Hirokawa, N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell 2003, 114, 229–239.
- Garnham, C.P.; Roll-Mecak, A. The chemical complexity of cellular microtubules: Tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton 2012, 69, 442–463.
- Halpain, S.; Dehmelt, L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006, 7, 1–7.
- Lee, M.K.; Cleveland, D. Neuronal Intermediate Filaments. Annu. Rev. Neurosci. 1996, 19, 187–217.
- Sheng, L.; Leshchyns’Ka, I.; Sytnyk, V. Cell adhesion and intracellular calcium signaling in neurons. Cell Commun. Signal. 2013, 11, 94.
- Leshchyns’Ka, I.; Sytnyk, V. Reciprocal Interactions between Cell Adhesion Molecules of the Immunoglobulin Superfamily and the Cytoskeleton in Neurons. Front. Cell Dev. Biol. 2016, 4, 9.
- Waites, C.L.; Craig, A.M.; Garner, C. Mechanisms of Vertebrate Synaptogenesis. Annu. Rev. Neurosci. 2005, 28, 251–274.
- Stanika, R.; Campiglio, M.; Pinggera, A.; Lee, A.; Striessnig, J.; Flucher, B.E.; Obermair, G.J. Splice variants of the CaV1.3 L-type calcium channel regulate dendritic spine morphology. Sci. Rep. 2016, 6, 34528.
- Dillon, C.; Goda, Y. The Actin Cytoskeleton: Integrating Form and Function at the Synapse. Annu. Rev. Neurosci. 2005, 28, 25–55.
- Eyang, Y.; Ecalakos, N. Presynaptic long-term plasticity. Front. Synaptic Neurosci. 2013, 5, 8.
- Sheng, L.; Leshchyns’Ka, I.; Sytnyk, V. Neural Cell Adhesion Molecule 2 (NCAM2)-Induced c-Src-Dependent Propagation of Submembrane Ca2+ Spikes Along Dendrites Inhibits Synapse Maturation. Cereb. Cortex 2019, 29, 1439–1459.
- Hussman, J.P.; Chung, R.-H.; Griswold, A.J.; Jaworski, J.M.; Salyakina, D.; Ma, D.; Konidari, I.; Whitehead, P.L.; Vance, J.M.; Martin, E.R.; et al. A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol. Autism 2011, 2, 1.
- Molloy, C.A.; Keddache, M.; Martin, L.J. Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Mol. Psychiatry 2005, 10, 741–746.
- Scholz, C.; Steinemann, D.; Mälzer, M.; Roy, M.; Arslan-Kirchner, M.; Illig, T.; Schmidtke, J.; Stuhrmann, M. NCAM2 deletion in a boy with macrocephaly and autism: Cause, association or predisposition? Eur. J. Med. Genet. 2016, 59, 493–498.
- Winther, M.; Berezin, V.; Walmod, P.S. NCAM2/OCAM/RNCAM: Cell adhesion molecule with a role in neuronal com-partmentalization. Int. J. Biochem. Cell Biol. 2012, 44, 441–446.
- Petit, F.; Plessis, G.; DeCamp, M.; Cuisset, J.-M.; Blyth, M.; Pendlebury, M.; Andrieux, J. 21q21 deletion involving NCAM2: Report of 3 cases with neurodevelopmental disorders. Eur. J. Med. Genet. 2015, 58, 44–46.
- Han, M.-R.; Schellenberg, G.D.; Wang, L.-S.; Initiative, T.A.D.N. Genome-wide association reveals genetic effects on human Aβ 42 and τ protein levels in cerebrospinal fluids: A case control study. BMC Neurol. 2010, 10, 90.
- Kimura, R.; Kamino, K.; Yamamoto, M.; Nuripa, A.; Kida, T.; Kazui, H.; Hashimoto, R.; Tanaka, T.; Kudo, T.; Yamagata, H.; et al. The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between β-amyloid production and tau phosphorylation in Alzheimer disease. Hum. Mol. Genet. 2007, 16, 15–23.
- Niederhoffer, K.Y.; Fahiminiya, S.; Eydoux, P.; Mawson, J.; Nishimura, G.; Jerome-Majewska, L.A.; Patel, M.S. Diagnosis of Van den Ende-Gupta syndrome: Approach to the Marden-Walker-like spectrum of disorders. Am. J. Med. Genet. Part A 2016, 170, 2310–2321.
- Griesi-Oliveira, K.; Suzuki, A.M.; Alves, A.Y.; Mafra, A.C.C.N.; Yamamoto, G.L.; Ezquina, S.; Magalhães, Y.T.; Forti, F.L.; Sertie, A.L.; Zachi, E.C.; et al. Actin cytoskeleton dynamics in stem cells from autistic individuals. Sci. Rep. 2018, 8, 1–10.
- Torrico, B.; Antón-Galindo, E.; Fernàndez-Castillo, N.; Rojo-Francàs, E.; Ghorbani, S.; Pineda-Cirera, L.; Hervás, A.; Rueda, I.; Moreno, E.; Fullerton, J.M.; et al. Involvement of the 14-3-3 Gene Family in Autism Spectrum Disorder and Schizophrenia: Genetics, Transcriptomics and Functional Analyses. J. Clin. Med. 2020, 9, 1851.
- Mukaetova-Ladinska, E.B.; Arnold, H.; Jaros, E.; Perry, R.; Perry, E. Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol. Appl. Neurobiol. 2004, 30, 615–623.
- Pham, E.; Crews, L.; Ubhi, K.; Hansen, L.; Adame, A.; Cartier, A.; Salmon, D.; Galasko, D.; Michael, S.; Savas, J.N.; et al. Progressive accumulation of amyloid-β oligomers in Alzheimer’s disease and in amyloid precursor protein transgenic mice is accompanied by selective alterations in synaptic scaffold proteins. FEBS J. 2010, 277, 3051–3067.
- Penzes, P.; VanLeeuwen, J.-E. Impaired regulation of synaptic actin cytoskeleton in Alzheimer’s disease. Brain Res. Rev. 2011, 67, 184–192.
- Scheff, S.W.; Price, D.A. Synaptic pathology in Alzheimer’s disease: A review of ultrastructural studies. Neurobiol. Aging 2003, 24, 1029–1046.
- Selkoe, D.J. Alzheimer’s Disease Is a Synaptic Failure. Science 2002, 298, 789–791.
- Smith, K.R.; Damiano, J.; Franceschetti, S.; Carpenter, S.; Canafoglia, L.; Morbin, M.; Rossi, G.; Pareyson, D.; Mole, S.; Staropoli, J.F.; et al. Strikingly Different Clinicopathological Phenotypes Determined by Progranulin-Mutation Dosage. Am. J. Hum. Genet. 2012, 90, 1102–1107.
