Pyrrolnitrin (PRN) is a microbial pyrrole halometabolite of immense antimicrobial significance for agricultural, pharmaceutical and industrial implications. The compound and its derivatives have been isolated from rhizospheric fluorescent or non-fluorescent pseudomonads, Serratia and Burkholderia. They are known to confer biological control against a wide range of phytopathogenic fungi, and thus offer strong plant protection prospects against soil and seed-borne phytopathogenic diseases. Although chemical synthesis of PRN has been obtained using different steps, microbial production is still the most useful option for producing this metabolite. In many of the plant-associated isolates of Serratia and Burkholderia, production of PRN is dependent on the quorum-sensing regulation that usually involves N-acylhomoserine lactone (AHL) autoinducer signals. When applied on the organisms as antimicrobial agent, the molecule impedes synthesis of key biomolecules (DNA, RNA and protein), uncouples with oxidative phosphorylation, inhibits mitotic division and hampers several biological mechanisms.
1. Introduction
Of the 5–30 million species on the Earth, fewer than 2 million have been described and fewer than 1% have been explored for a vast repertoire of new natural products with socio-economic significance. Hence, it is reasonable to expect that many more natural products not only from known species, but also from unidentified organisms are yet to come to benefit humanity and the environment. Natural products offer unique structural molecules unparalleled by any other molecular family with an array of biological activities such as for drug leads. The many natural products that occupy the market today without any chemical modification are a testimony to the remarkable properties of secondary molecules produced by an array of plants, insects, animals, microbes and numerous species of marine organisms.
Secondary metabolites are small heterogenous organic molecules that display prominent ecological benefits to the host organisms in providing defense against predators, parasites, diseases, interspecies nutritional competence, and competitive edge over interaction with the environment. Extensive microbial structural diversification has led to maximizing chemical diversity in terms of the secondary metabolite resources that triggered scope for new drug leads. Since natural products have reflected a wide array of therapeutic and biological applications (antibiotic, anti-inflammatory, antimicrobial, antitumor, anticancer, antiparasitic and immunosuppressing agents as well as enzyme inhibitors), the scope for further exploration of uncharacterized molecules of plant and microbial origin has always remained a focused area for identifying new leads for pharmaceutical and agro-chemical usages.
Continuously changing environmental patterns, the emergence of new diseases and resurgence of resistance towards existing drugs have led to an extensive search for novel natural metabolites at a rapid rate. Low molecular size secondary metabolites from living entities have been obtained with typical therapeutic, biological and agricultural implications including antimicrobials, growth promoters, disease suppressers, enzyme inhibitors, health stimulators, and biocontrol agents against pathogenic fungi, bacteria and insects. Systematic strategies for obtaining bioactive metabolites include isolation and identification of known secondary metabolites with biological activities unmatched with the molecular libraries or search for unknown natural molecules with versatile bioactivities. For both these options, the microbial world offers a great repository of natural molecules due to their extensive chemical diversity. However, there remains limitations of the culturability of microbial species and the expression of desired molecular traits or chemical species under isolated culture conditions. To overcome this, metagenomics has emerged to represent vast structural diversity of taxonomic communities with multi-functionalities having diverse chemical structures and functions.
Thus, secondary metabolites are supposed to be conserved in the species, evolved in a competitive environment, emerged to serve purposes other than primary metabolism, secreted for specific physiological or defense-related reasons, related with the habitat of producing organisms, blessed with complex chemical structures and clubbed with diverse bioactivity. These attributes potentiate the usefulness of structurally diverse but functionally sound microbial biomolecules in therapeutics, and agricultural, industrial and environmental applications. We discuss structural, chemical, biological and functional perspectives of one of the earliest known pyrrole antibiotic antifungal metabolite of microbial origin, pyrrolnitrin, which has witnessed laboratory to commercial implications.
2. Pyrrolnitrin (PRN)
Pyrrolnitrin [3-chloro-4-(2-nitro-3-chlorophenyl) pyrrole] is a phenylpyrrole derivative containing two chlorine atoms and a nitro group. PRN, isolated from
Pseudomonas pyrrocinia and various other pseudomonads, was classified as halometabolite in as early as 1964. Later, the compound was biosynthesized using tryptophan as supplement in the medium and chemically synthesized by Nakano et al. Biosynthesis of PRN in
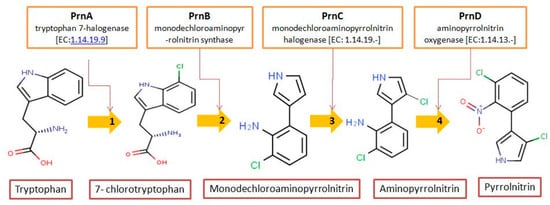
Pseudomonas aureofaciens ATCC 15926 has shown that L-tryptophan is a direct precursor (
Figure 1). However, high yield of PRN was obtained in D-tryptophan amended medium. Tryptophan analogs amended in the fermentation medium can also yield a series of PRN-like derivatives (
Table 1) with low antimicrobial activity than the native parent compound.
Figure 1. Biosynthetic steps in the synthesis of pyrrolnitrin. 7-chlorotryptophan is formed from tryptophan due to flavin-dependent halogenation catalyzed by the enzyme tryptophan 7-halogenase (PrnA). Further, the enzyme PrnB (monodechloroaminopyrrolnitrin synthase catalyzes formation of monodechloroaminopyrrolnitrin from 7-chlorotryptophan while the enzyme PrnC leads to catalytic reaction for the conversion of monodechloroaminopyrrolnitrin into aminopyrrolnitrin. In the last step, aminopyrrolnitrin is converted to pyrrolnitrin with the help of the enzyme PrnD (aminopyrrolnitrin oxygenase).
Table 1. Derivatives of pyrrolnitrin biosynthesized by Pseudomonas aureofaciens
| IUPAC Name |
Common Name |
Structure |
Extinction Coefficient ƛmax MeOH (log ε) |
Molecular Formula |
| Molar Mass/ Molecular Weight |
| 3-(2-amino-3-chlorophenyl)-pyrrole |
Mono-chloro-amino-pyrrolnitrin (MCA) |
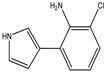 |
- |
303 (3.52) |
C10H9ClN2 |
Exact Mass: 192.05
Mol. Wt.: |
Burkholderia cepacia complex, JKB9, showing broad-spectrum antifungal activity, was held responsible for suppressing growth of
Phytophthora capsici, Fusarium oxysporum and
Rhizoctonia solani. This strain, which has shown stronger antifungal activity than
Burkholderia strains KCTC2973 and ATCC25416 against
Phytophthora blight, was confirmed for PRN production using thin layer chromatography (TPC), high performance liquid chromatography (HPLC) and Nuclear Magnetic Resonance (NMR) spectrometric studies. Complete genome sequencing of
Burkholderia pyrrocinia 2327
T revealed insights into the cells possessing antibiotic capabilities for the biosynthesis of PRN. Cloning of gene clusters responsible for encoding enzymes involved in the production of pyrrolnitrin in organisms has greatly helped in marking of the biosynthetic routes. Using an antibiotic producing strain of
P. fluorescens cloned four gene clusters to elucidate biochemistry of these molecules and to link it with the enzymes that may offer the routes for the synthesis of new chemical structures. Earlier,
prnABCD operon from
P. protegens Pf-5 was co-expressed in tomato plants with universal vector IL-60 and successfully demonstrated resistance to damping-off disease caused by
R. solani .
Microbial wild type strains secrete PRN in low quantity (
Table 2) and production varies with the medium constituents.
P. aureofaciens ATCC 15926 strain when grown in minimal medium, secreted PRN in low concentration (<0.3 µg mL
−1). Even optimized variation of constituents in growth medium could not increase PRN production. However, the production enhanced by 30-fold when
P. aureofaciens ATCC 15926 was mutated with N-methyl-N’-nitro-N-nitrosoguanidine. Addition of DL-tryptophan (1 mg mL
−1) in CMM medium also doubled PRN production after 120 h but additional amount of tryptophan resulted in less yield.
Table 2. Characteristics of PRN production by different microbial species inhabiting several ecohabitats
| Sr. no. |
Producer |
Habitat |
Medium |
Physical Condition |
Incubation Period (Days) |
Concentration |
Significance |
|
| 1. |
Pseudomonas pyrrocinia |
- |
Bouillon Medium |
- |
- | 192.64 |
| ND |
Antibiotic, antifungal nature |
|
| Silica gel G |
35 cm × 1.5 cm |
Chloroform: methanol (9:1) |
- |
|
| Silica gel (40 μm) |
35.6 cm × 1.75 cm |
Benzene: hexane (2:1); Benzene; Benzene: acetone (1:1); Acetone; methanol |
TLC - bioautography |
|
H
6O
2N
2Cl
2 and molecular weight 257.07 gmol
−1. The melting point of PRN, which is sparingly soluble in water, petroleum ether and cyclohexane, but more soluble in ethanol, butanol, ethyl acetate, ethyl ether, snf chloroform, is 124.5 °C. Elemental analysis of the compound reflected C, 46.71%; H, 2.33%; O, 12.45%; N, 10.89%; and Cl, 27.68%. PRN separation was achieved by different methods like chromatography TLC and HPLC while structural features have been elucidated using Fourier transform infrared spectroscopy (FTIR), nuclear magnetic spectroscopy (NMR
1H and
13C) and mass spectroscopy (LC-MS and GC-MS).
Separation of PRN from bacterial media extract using TLC utilized various stationary phases such as silica gel G, GF
254, 60 F
254, KCI8 F, C18 Glass and several mobile phases. PRN can be detected on TLC under UV transilluminator and visualized by spraying diazotized sulfanilic acid (DSA) or Pauly’s, Ehrlich’s and van Urk’s reagent to develop maroon and violet color, respectively or H
2SO
4 on Silica Gel G plate. The R
f value for various TLC system served to identify PRN from different bacterial species. The compound has been analyzed by retention time in gradient HPLC system but isocratic solvent system of 45% water, 30% acetonitrile, and 25% methanol also separated pyrrolnitrin at 252 nm in preparative HPLC. Modifications in the polarity of solvents, mobile-stationary phase and elution methods are key strategies to quantify PRN using HPLC (
Table 4). Yellow colored PRN molecule isolated from
P. pyrrocinia absorbs at 252 nm with molar extinction coefficient of ε = 7500 in ethanol. Myxobacterial PRN also showed λ
max at 252 nm in methanol. Functional group stretching in FTIR vary with different PRN derivatives due to its structural features. Typical bond stretching at 1530 and 1375 cm
−1 characterized for nitro group while 3489 cm
−1 represent pyrrole ring. Similarly, PRN isolated from supernatant of fermented medium inoculated by
Myxococcus fulvus strain Mx f147 indicated infrared spectrum to confirm pyrrole ring (3460), nitro group (1530 and 1375), CH
3 (stretch) (1460) and C=C aromatic weak intensity (1600). Mass spectroscopy (MS) of PRN is ascertain using different ionization techniques. MS of PRN isolated from
Pseudomonas cepacia B37w showed molecular ion at
m/z 256 with the formula C
10H
6C
12N
2O
2. Electrospray mass spectroscopy (negative ion spectrum) of PRN further confirmed (mass-to-charge ratio;
m/z) at 256. High-resolution mass spectrometry of the two molecular ions gave
m/z 255.9826 and 257.9777, respectively, indicating the molecular formula C
10H
6N
2O
235C
12 and C
10H
6N
2O
235Cl
37Cl.
Table 4. Several HPLC methods adopted to separate and quantify pyrrolnitrin from microbes using different solvent system.
| Column |
Flow Rate (mL/min−1) |
Solvent System |
Detector |
Retention Time (min) |
|
| RP 18 |
2 |
Methanol: water (70:30; v/v) |
- |
- |
|
| 3-chloro-4(2-amino-3-chlorophenyl)-pyrrole |
Di-chloro-amino (DCA)
(amino-pyrrolnitrin) |
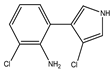 |
212 (4.46) |
302 (3.57) |
C10H8Cl2N2 |
Exact Mass: 226.01
Mol. Wt.: 227.09 |
| 2. |
P. aureofaciens, P. fluorescens, P. multivorans |
- |
CMM, Synthetic C, E |
27 °C, shaker |
7 |
0.32–126 (µg mL−1) |
PRN widespread in groups of Pseudomonas |
|
| 50 mm × 4.6 mm I.D. guard |
1.0 |
Acetonitrile: methanol: water (1:1:1) |
UV (254 nm) |
10 |
2, 3 dichloro-4-(2-amino-3-chlorophenyl)-pyrrole |
Tri-chloro-amino
(TCA) |
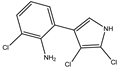 |
212 (4.54) |
| |
3. |
P. aureofaciens |
Silica gel (60 μm) | - |
- |
Chloroform: hexane (1:1, 1.5:1, 2:1, 5:1) (v/v); chloroform; chloroform-acetone (5:1, 1:1) (v/ | 302 (3.61) |
v); acetone |
Bioassay with R. solani |
|
| Rainin Dynamax C18 (21.4 × 250 mm) |
- |
Acetonitrile: water (3:2; v/v | C | 10 | H7Cl3N2 |
Exact Mass: 259.97
Mol. Wt.: 261.53 |
| CMM |
27 °C, shaker |
) fractions collected at 9.5 to 12.5 min and re-chromatographed on a silica column eluted with chloroform: hexane (1:1; | 5 |
v/v | 9.5 to 50 (µg mL | −1) |
Production of substituted PRN from Tryptophan analogs |
) |
- | |
13.5 |
|
3-chloro-4-(3-chloro-2nitro-phenyl)-1H pyrrole |
Pyrrolnitrin (PRN) |
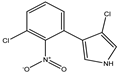 |
212 (4.39) |
252 (3.83) |
C10H |
| 4. |
P. aureofaciens |
Sephadex LH-20 |
- |
Methanol |
pHPLC |
|
| C-18 column, 5 µm |
- |
Isocratic acetonitrile: methanol: water (1:1:1) | 6 |
- |
CMM | Cl |
30 °C, shaker | 2N2O2 |
- |
- |
| Exact Mass: |
5 | 255.98
Mol. Wt.: 257.07 |
| 18.35–19.9 (µm) |
Possible pathway discussed |
|
2, 3 dichloro-4-(2-nitro-3-chlorophenyl) pyrrole |
2-chloro-pyrrolnitrin
(2-CPRN) |
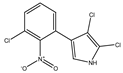 |
212 (4.47) |
240
280 |
C10H5Cl3N2O2 |
Exact Mass: 289.94
Mol. Wt.: |
| 5. | 291.52 |
| Pseudomonas cepacia | B37w (NRRL B-14858) |
Rhizosphere |
Sabouraud Maltose Broth |
- |
6 |
2.133 (mg L−1) |
Efficacy against F. Sambucinum incited potato dry rot disease |
|
Silica gel 60 (0.015–0.040 mm; Merck) |
- |
Dichloromethane then methanol |
TLC |
|
| Hypersil octyldecyl saline (2.1 mm diameter by 10 cm) |
- |
Water: methanol from 0%: 100 % and gradually changing up to 100%: 0% |
- |
between 10-15 |
|
2-(2-Heptenyl)-3-methyl-4(1H) quinolone |
- |
| 6. |
Pseudomonas cepacia | 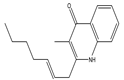 |
LT4-12- W | - |
- |
C17H21NO |
Exact Mass: 255.16
Mol. Wt.: 255.35 |
| Apple leaves |
Silica gel (H60) |
- |
Dichloromethane | Mineral Salt, Nutrient Broth, Kings medium B |
Bioautography |
|
| Reverse phase 18 |
0.7 |
0 min 50% methanol in water
15 min 100 % methanol in water
17 min 100% methanol in water
| 27 °C, 200 rpm |
20 min 50% methanol in water | 7 |
25 min 50% methanol in water | 1) MS: 51.50 (mg L−1) 2) NB: 7.20 (mg L−1) 3) KMB: 5.50 (mg L−1) |
Production of phenylpyrrole metabolites with respect to time |
|
UV (252 nm) |
15.4 |
|
2,3-dichloro-4-(2-nitrophenyl) pyrrole |
Iso-pyrrolnitrin |
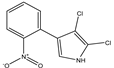 |
- |
- |
Silica gel | C10H6Cl2N2O2 |
| 7. |
B. cepacian |
- |
(20 × 170 mm, Wakogel C-200) | Exact Mass: | 255.98 |
Mineral Salt |
Mol. Wt.: 257.07 |
| 27 °C, shaker |
7 |
Benzene, 10% ethyl acetatobenzene, 20% ethyl acetate benzene and finally ethyl acetate | ND |
Delays postharvest fruit rot in strawberries |
TLC |
|
| C-18 reverse phase (125 × 4.6 mm) |
- |
Methanol: water (70:30; v/v) |
UV (252 nm) |
- |
| |
3-chloro-4-(2-nitro-3-chloro-6-hydroxyphenyl) pyrrole |
Oxy-pyrrolnitrin |
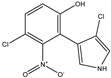 |
- |
- |
C10H6Cl2N2O3 |
Exact Mass: 271.98
Mol. Wt.: 273.07 |
Structurally, PRN possesses benzene and pyrrole rings with chlorine atoms on both of them and nitro and chlorine units to form an unusual natural skeleton. It has chlorine moiety to contribute more towards biological activity in comparison to its bromine derivative. Consequently, several natural congeners of PRN such as amino-pyrrolnitrin, iso-pyrrolnitrin, 2-chloropyrrolnitrin, oxy-pyrrolnitrin, 4-fluoropyrrolnitrin, and 3-fluoro-3-dechloropyrrolnitrin have been reported. Brominated derivatives of PRN can be synthesized by replacing chlorine ion with bromine in the presence of sodium bromide.
2.1. Pyrrolnitrin: Chemical Synthesis
PRN is positive towards Ehrlich’s reagent where pyrrole ring gets condensed with p-dimethylaminobenzaldehyde to form the violet color complex. Pauly’s coupling reaction yields red color and gives a negative reaction to the ferric chloride nitro group detection test. PRN can be oxidized by chromic acid to form corresponding compound which on oxidation with permanganate, yields carboxylic acid.
Modern synthetic targets for chemical synthesis require regiospecific polysubstituted aromatic or heteroaromatic components . PRN is chemically synthesized by α-block of pyrrole ring and 2-nitro-3-chloroacetophenone, and subsequent chlorination at 4-position and oxidation of methyles using sulfurilchloride followed by decarboxylation. In another approach, 2-Methyl-4-(2-nitro-3-chloro-phenyl)-5-ethoxycarbonyl-pyrrole was prepared in various steps from 2-amino-3-chloro-toluene. One of the most versatile synthetic approach for PRN allowed access to analog compounds such as monodechloroaminopyrrolnitrin and aminopyrrolnitrin. This step facilitated PRN synthesis using Suzuki–Miyaura cross-coupling of an appropriately halogenated pyrrole pinacolboronate ester with halogenated arylpyrroles using 2,6-disubstituted nitrobenzenes or 2,6-disubstituted anilines. Palladium-catalyzed coupling of 1-(triisopropylsilyl-3-substituted pyrroles with arylhaildes has also been described. However, chemical synthesis of PRN makes synthetic route cost extensive and pose threat to the environment. Furthermore, the chemical process utilizes noxious chemicals, high temperature and pressure, more energy and yield poor regioselectivity with lack of public acceptability. Thus, chemical industries prefer microbial species for more selective, greener and cost-effective approach for synthesizing PRN.
2.2. Microbial Pyrrolnitrin Production and Recovery
Microbial synthesis of PRN is easy, reliable and eco-friendly and requires low-cost medium constituents, ambient conditions for growth and production, the least additional energy requirements and minimum expensive equipment. This is the major reason microbial synthesis of PRN has become the preferred alternative to chemical processes. After initial isolation of PRN from
Pseudomonas pyrrocinia and thereafter reports from different fluorescent and non-fluorescent
Pseudomonas species, several strains of
Burkholderia cepacia,
Corallococcus exiguus, Cystobacter ferrugineus, Enterobacter agglomerans, Myxococcus fulvus, Serratia spp. and
Actinosporangium vitaminophilum have been classified to produce PRN in varying quantities.
Serratia plymuthica and
S. ruhidaea are identified for enhanced production of PRN. Recently, a strain belonging to
| 8. |
| Enterobacter agglomerans |
| IC1270 |
| Grapes rhizosphere |
| Potato Dextrose Agar |
Incubated on agar plate |
5 |
ND |
Possible role of a combination of Chitinases and pyrrolnitrin in antagonism |
|
| Silicic acid |
(240 × 22 mm) |
Diethyl ether and methanol |
- |
| - | |
| 1.0 |
2-min initialization at 10% ACN: 0.1% TFA; 20-min linear gradient to 100% ACN: 0.1% TFA |
990-photodiode array detector |
- |
|
9. |
B. cepacia NB-1 |
Ponds in botanical garden of Tubingen, Germany |
Minimal medium |
27 °C, aeration rate 0·5 vvm, stirrer speed 150 rev min−1, pH −7.0 |
5 |
0.54 (mg L−1) |
- |
RP C-18 flash | PRN block ETS |
Water and methanol | Neurospora crassa 74 A; inhibition of Streptomycine spp. |
|
| TLC |
|
10. |
Burkholderia cepacia 5.5B (ATCC 55344) Wild Type |
Soil sample, North Carolina |
| Nucleosil C-18 |
|
Acetonitrile: water (20 to 100%) |
- |
27.5 |
|
Nutrient broth, Mineral salt |
25 °C, at 200 rpm, pH 5.8 |
- |
RP C-18 (MPLC) | 5 |
50% to 100% aq methanol |
HPLC |
|
| RP C-18 column |
1.0 | NB: 35.59; MS: 28.54 (mg 10 | 12 | cfu) |
Biocontrol of | Rhizoctonia | stem rot of poinsettia |
Isocratically 45% water: 30% acetonitrile: 25% methanol | |
| - |
- |
|
11. |
Pseudomonas fluorescens psd |
Roots of Vigna mungo |
Standard succinate medium (SSM) |
- |
- |
ND |
Biocontrol property of plants protected from strain |
|
Silica gel (60 μm) |
- |
Toluene |
- |
|
| C-18 RP column |
|
10% acetonitrile: water (v/v) (both acidified with 0.1% amino acid) run for 2min linear gradient 100% acetonitrile (acidified with 0.1% amino acid) |
- |
18 |
|
12. |
Pseudomonas chlororaphis O6 |
- |
Nutrient broth, Mung bean medium |
28 °C 200 rpm |
- |
1.7 (µg mL−1) |
Regulation by glucose of PRN production influenced biocontrol of tomato leaf blight |
|
| - |
- |
30 ~ 60% aq acetonitrile (for 70 min) |
- |
68.9 |
|
13. |
Acinetobacter haemolyticus A19 |
Wheat rhizosphere |
Luria broth |
- |
- |
|
NMR spectroscopy is widely used for analytical measurement of microbial metabolites. The PRN is confirmed by NMR spectrum with values: (i) 1H NMR: H-2 and H-5: 6.82 (m, 2H); H-5′: 7.41-7.53 (m, 3H); NH: 8.29 (br s, 1H); and (ii) 13C NMR δ value: 111.9 indicated C-3, 115.4 for C-4, 116.6 for C-5, 117.4 meant for C-2, while 124.8, 127.6, 128.6, 130.1, and 130.3 designated for C-3′, C-1′, C-6′, C-4′ and C-5′, respectively. Chemical shift (δ) values at 6.81 (m, 2H) indicate the presence of H-2 and H-5, 7.41: H-6′, 7.43: H-5′, 7.52: H-4′, 8.38: NH. NMR spectrum of purified PRN secreted by plasmid-mediated
A. haemolyticus A19 revealed the values δ: 6.2–6.6 (m, 2H, H-2, H-5), 6.77 (q 1H, H6), 7.03 (m, 1H, H-4), 7.38 (m2H, Ha, Hc) compared with standard 1H NMR spectrum of. PRN synthesized from
Myxococcus fulvus strain Mx f147 showed 13C NMR spectrum (in acetone-
d6; Bruker 400 MHz). Structural investigation of PRN with X-ray analysis revealed the presence of two molecules with observed density of 1.74 g/cm
3 that lie opposite to each other about the center of symmetry. It further confirmed the location of two Cl atoms in the asymmetric unit with 3D Patterson function, dihedral angle of the pyrrole, the benzene rings and chlorine substitution on pyrrole ring located apart from the nitro group.
2.4. Biochemistry of Pyrrolnitrin
Microbial synthesis of PRN requires D-tryptophan, but cost of precursor amino acid and intracellular secretion limits its large-scale production. The NO
2 group is derived from anthranilic acid, phenylalanine and tryptophan that could serve as a precursor for PRN secretion. However, anthranilic acid and L-phenylalanine usually decrease PRN secretion in
P. aureofaciens and
B. cepacian, while tryptophan stimulates PRN production. In the medium, L-tryptophan gets quick intake within the cells than the D- isomer but addition of L-isomer could not yield more PRN secretion. In actinomycetes, D-tryptophan enhances secretion of PRN when added separately in the culture medium and maximum accumulation was observed at stationary phase after 120 h. It indicated that the L-isomer of tryptophan enter cells quickly and participate in the protein synthesis, while D-tryptophan enter slowly and available at the time of antibiotic secretion. Besides, L-glutamic acid amended medium showed maximum antifungal activity, which substantially declined with the addition of L-tryptophan, L-valine, L-serine, L-phenylalanine and L-cysteine. In brief, D-tryptophan and L-glutamic acid are more direct precursors of PRN than any other amino acids.
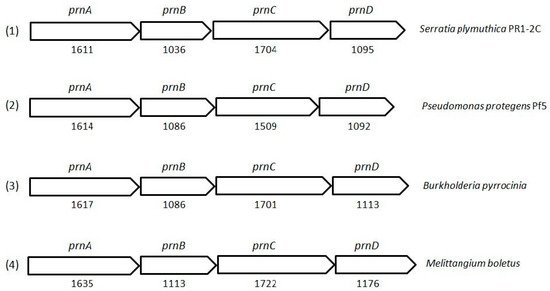
PRN biosynthesis was unraveled in
P. aureofaciens. Later, genes (
prnABCD operon) and corresponding enzymes involved were delineated in
P. fluorescens BL915 (
Figure 2). The biosynthesis of PRN occurs in four sequential steps: chlorination by prnA, rearrangement and decarboxylation by prnB, chlorination by prnC and oxidation by prnD enzyme (
Figure 1). This involves regioselective halogenation of tryptophan through the addition of chlorine into D-tryptophan by tryptophan 7-halogenase (prnA) following nucleophilic and electrophilic reactions and activation of intermediate lysine-chloramine species as the first step. Further, the reaction catalyzed by prnB shows structural similarity with two-domain indoleamine 2,3-dioxygenase enzyme (IDO) and involves several intermediary steps. The second step forms a binary complex that combines with L-tryptophan or 7-Cl-L-tryptophan to create a ternary complex. The third step in PRN biosynthetic pathway of
P. fluorescens leads to catalytic conversion of mono-chloro-deamino-pyrrolnitrin into amino-pyrrolnitrin by regioselectivity using halogenating and chlorinating enzyme. In the last step, prnD catalyzes the oxidation of amino group of aminopyrrolnitrin to nitro group and thus forms PRN.
Figure 2. Cluster organization of pyrrolnitrin biosynthetic genes in S. plymuthica PR1-2C (1), P. protegens Pf5 (2), B. pyrrocinia (3) and myxobacterium Melittangium boletus (4). Nucleotide sequence (NT size indicated for prnA–D genes) of the bacterial species was obtained from KEGG database.
Aminopyrrolnitrin oxidase or arylamine oxygenase (rieske N-oxygenase) catalyzes oxidation of an arylamine into the arylnitro group. Except prnB, trptophan-7-halogenase (prnA), monodechloroaminopyrrolnitrin (prnC) and aminopyrrolnitrin oxidase (prnD) enzymes require flavin reductase (
prnF) gene located close to the
prnABCD operon which is considered as a part of the cluster. Bioinformatics clubbed with the biochemical tools identified the role of
prnF gene in
prnD-catalyzed unusual arylamine oxidation in
P. fluorescens Pf-5. The
prnF and
prnD genes form a two-component oxygenase system, in which the gene product enzyme prnF supplies the reduced flavin to prnD. The prnF requires NADH as an electron donor to reduce FAD so that reduced FAD supplies electrons from NADPH to the prnD oxygenase component through protein-protein interactions in order to protect the flavin from oxidation.
The
prnF gene having molecular mass of 17kD with GC content of 62%, encodes for a polypeptide chain of 160 amino acids. The enzyme belongs to flavin:NAD(P)H reductases family with part of two-component monooxygenase systems and its C-terminal region possesses highly conserved GDH motif for NAD(P)H binding]. It resembles with PheA2, SnaC, VlmR, ActVB and HpaC with 31.5%, 28.6%, 26.4%, 25.6%, and 25.5% amino acid identity, respectively.
2.5. Pyrrolnitrin Derivatives
Several halogen variations of the PRN molecule have been isolated in the past in the form of bromo-analogs of pyrrolnitrin from fermentation of
P. aureofaciens in sodium bromide with low antifungal activity. In addition, 2-chloropyrrolnitrin contain an additional chlorine atom which possesses about 10% of the antifungal activity of PRN. The pyrrolomycin (B, C, D, E, F
1, F
2a, F
2b, and F
3) derivatives encompass a chlorine or bromine atom at the 3-position of the pyrrole ring, and either two chlorine atoms at positions 4 and 5 or one chlorine and one bromine at any of these positions have shown significant antifungal activity. Novel oxidized derivatives of pyrrolnitrin including two new pyrrolnitrin analogs, namely 3-chloro-4-(3-chloro-2-nitrophenyl)-5-methoxy-3-pyrrolin-2-oneand4-chloro-3-(3-chloro-2-nitrophenyl)-5-methoxy-3-pyrrolin-2-onehave, were reported from
B. cepacia K87.
Furthermore, number of
de-chloro and
de-nitro derivatives of PRN and the isomers were synthesized by cyclization of enamine reaction, hydrolysis, carboxylation and Mannich’s reaction. The strongest antifungal activity of PRN and its analogs resulted when it got unsubstituted by ester group at any position. The antifungal activity become more stronger when shift of NO
2 group was increased. Few PRN derivatives such as denitropyrrolnitrin (3-chloro-4-(3-chlorophenyl), bromo analog: 3-chloro-4-(3-bromophenyl)pyrrole) and trifluoromethyl derivative (3-chloro-4-(3-trifiuorornethyl) pyrrole) were strong antimicrobials. However, among all the analogs homologous to NO
2 group of pyrrolnitrin, PRN has remained the strongest biologically active compound. The UV irradiation of 2-(pyrrol-3-yl)nitrobenzene moiety of PRN in an anhydrous aprotic solvent yielded 7,4′-dichlorospiro(1,3-dihydrobenzo(c)isoxazole-3,3′-pyrrolin-2′-one) by the intramolecular oxidation. Hence, the photodegradation of PRN depends on aqueous reaction media and the nature of its excited state.
3. Applications of Pyrrolnitrin
3.1. Biological Activity
Structure–activity mechanism reveals that the primary target of PRN lies in the cell membrane to impede protein, RNA, DNA synthesis and uncouple the normal electron flow in the respiratory electron transport chain. The metabolite has demonstrated biological activity at low concentration and act as an uncoupler of oxidative phosphorylation in
Neurospora crassa. High concentration of PRN causes impairment of electron transport in flavin region and cytochrome c oxidase; accumulation of glycerol; synthesis of triacyl glycerol leading to leakage of cell membrane and inhibition of cell growth; in vitro activity against bacteria and fungi in the range of 1–100 µg mL
−1; in vitro activity against leukemia and melanoma cell lines; and moderate antimycobacterial activity at 8 µg mL
−1. The halometabolite was used as a drug lead for fenipoclonil and fludioxonil synthesis. The amino derivative of PRN was identified as an androgen receptor antagonist. PRN has the unique property to persist actively in the soil over a month, and can be readily diffused and slowly released after lysis of host bacterial cell. However, the compound is sensitive to decomposition due to light.
Inhibitory effect of PRN is seen on the mitochondrial electron transport system of
Neurospora crassa 74A. Studies using N,N,N’,N’-tetramethyl-p-phenylenediamine dihydrochloride (TMPD) confirmed that PRN block transfer of electron between the dehydrogenases and cytochrome c-oxidase components of the respiratory chain. At low concentrations, PRN uncouples oxidative phosphorylation in
Neurospora mitochondria and impedes electron transport in both the Flavin region and cytochrome C oxidase at high concentration. PRN also function as a signal molecule, beyond its role as a bioactive molecule to suppress fungal and affected cell motility. Antifungal activity of the compound increased at pH 6.0, became maximum at pH 10 or 11 and declined after pH 11. Temperature influence on antifungal activity was maximum at 28 °C. Similarly, 2% NaCl content in the medium showed maximum activity. Such studies indicated more scope for medium modifications for obtaining maximum PRN production followed by maximizing biological activity of the compound.
The quorum-sensing system related regulation of PRN is reported in a chitinolytic bacterium
S. plymuthica strain HRO-C48, that protects oilseed rape crop from
Verticillium wilt. The mutant deficient in PRN production shown the ability to produce the compound in the medium supplemented with chemically synthesized N-hexanoyl-HSL and N-3-oxo-hexanoyl-HSL (OHHL) (100 µM), thus suggesting that quorum sensing (QS) ability regulated PRN biosynthesis. While investigating the role of N-acylhomoserine lactone (AHL)-dependent quorum sensing for expressing antifungal traits, It was found that PRN expression was positively regulated by
CepR gene at transcription level. PRN is reported to have significant antimicrobial potential (
Table 5) against
Streptomyces antibioticus, S. violaceoruber, Paecilomyces variotii and
Penicillium puberulum. However,
S. prasinus, S. ramulosus, Aspergillus proliferans and
A. terreus showed tolerance to pyrrolnitrin. PRN displayed activity against
Ustilago maydis, Candida albicans, Hansenula anomala, Arthrobacter oxidans, Bacillus coagulans, B. lichenifernis, B. subtilis and
B. thuriengiensis at low concentration. Pyrrolnitrin produced by
Pseudomonas chlororaphis strain PA23 exhibited nematicidal and repellent activity against
Caenorhabditis elegans. Co-culturing
P. chlororaphis and
C. elegans enhanced expression of biocontrol-related
phzA, hcnA, phzR, phzl, rpoS and
gacS genes and, thus, contributed to the fast killing of nematode in bacterial interaction.
Table 5. Bioactivity spectrum of pyrrolnitrin against bacteria, fungi and nematodes.
| Sr. No. |
Name of Test Microorganism |
PRN (µg mL−1) |
|
| Bacteria |
| 1. |
Staphylococcus aureus 209-P |
6.2 |
|
| 2. |
Escherichia coli |
250 |
|
| 3. |
M. tuberculosis CIP 103471 |
4.0 |
|
| 4. |
M. avium CIP 103317 |
8.0 |
|
| 5. |
M. smegmatis CIP 103599 |
16.0 |
|
| 6. |
M. gordonae CIP 6427 |
>16.0 |
|
| 7. |
M. marinum CIP 6423 |
>16.0 |
|
| Yeast |
| 8. |
Candida albicans |
1.0 |
|
| 9. |
Saccharomyces cerevisiae |
10.0 |
|
| 10. |
Cryptococcus neoformans |
< 0.78 |
|
| - |
2 |
15 (mg L | −1 | ) |
Plasmid-mediated pyrrolnitrin production by | A. Haemolyticus | A19 |
|
| 14. |
Pseudomonas chlororaphis strain PA23 |
- |
M9 medium + 1 mm MgSO4 + 0.2% glucose |
- |
5 |
ND |
Nematicidal and repellent activity against Caenorhabditis elegans |
|
| 15. |
Serratia marcescens ETR17 |
Tea rhizosphere |
Semi-solid pigment producing media |
30 °C |
8 |
ND |
Effective reduction of root-rot disease tea plant on talc-based formulations; Plant growth promoting activity |
|