The recently emerged SARS-CoV-2 is the cause of the global health crisis of the coronavirus disease 2019 (COVID-19) pandemic. No evidence is yet available for CoV infection into hosts upon zoonotic disease outbreak, although the CoV epidemy resembles influenza viruses, which use sialic acid (SA). Currently, information on SARS-CoV-2 and its receptors is limited. O-acetylated SAs interact with the lectin-like spike glycoprotein of SARS CoV-2 for the initial attachment of viruses to enter into the host cells. SARS-CoV-2 hemagglutinin-esterase (HE) acts as the classical glycan-binding lectin and receptor-degrading enzyme. Most β-CoVs recognize 9-O-acetyl-SAs but switched to recognizing the 4-O-acetyl-SA form during evolution of CoVs. Type I HE is specific for the 9-O-Ac-SAs and type II HE is specific for 4-O-Ac-SAs. The SA-binding shift proceeds through quasi-synchronous adaptations of the SA-recognition sites of the lectin and esterase domains. The molecular switching of HE acquisition of 4-O-acetyl binding from 9-O-acetyl SA binding is caused by protein–carbohydrate interaction (PCI) or lectin–carbohydrate interaction (LCI). The HE gene was transmitted to a β-CoV lineage A progenitor by horizontal gene transfer from a 9-O-Ac-SA–specific HEF, as in influenza virus C/D. HE acquisition, and expansion takes place by cross-species transmission over HE evolution. This reflects viral evolutionary adaptation to host SA-containing glycans. Therefore, CoV HE receptor switching precedes virus evolution driven by the SA-glycan diversity of the hosts. The PCI or LCI stereochemistry potentiates the SA–ligand switch by a simple conformational …
- COVID-19 Sialic acid HE
1. History
1. DefinitionThe recently emerged SARS-CoV-2 is the cause of the global health crisis of the coronavirus disease 2019 (COVID-19) pandemic. No evidence is yet available for CoV infection into hosts upon zoonotic disease outbreak, although the CoV epidemy resembles influenza viruses, which use sialic acid (SA). Currently, information on SARS-CoV-2 and its receptors is limited. O-acetylated SAs interact with the lectin-like spike glycoprotein of SARS CoV-2 for the initial attachment of viruses to enter into the host cells. SARS-CoV-2 hemagglutinin-esterase (HE) acts as the classical glycan-binding lectin and receptor-degrading enzyme. Most β-CoVs recognize 9-O-acetyl-SAs but switched to recognizing the 4-O-acetyl-SA form during evolution of CoVs. Type I HE is specific for the 9-O-Ac-SAs and type II HE is specific for 4-O-Ac-SAs. The SA-binding shift proceeds through quasi-synchronous adaptations of the SA-recognition sites of the lectin and esterase domains. The molecular switching of HE acquisition of 4-O-acetyl binding from 9-O-acetyl SA binding is caused by protein–carbohydrate interaction (PCI) or lectin–carbohydrate interaction (LCI). The HE gene was transmitted to a β-CoV lineage A progenitor by horizontal gene transfer from a 9-O-Ac-SA–specific HEF, as in influenza virus C/D. HE acquisition, and expansion takes place by cross-species transmission over HE evolution. This reflects viral evolutionary adaptation to host SA-containing glycans. Therefore, CoV HE receptor switching precedes virus evolution driven by the SA-glycan diversity of the hosts. The PCI or LCI stereochemistry potentiates the SA–ligand switch by a simple conformational shift of the lectin and esterase domains. Therefore, examination of new emerging viruses can lead to better understanding of virus evolution toward transitional host tropism. A clear example of HE gene transfer is found in the BCoV HE, which prefers 7,9-di-O-Ac-SAs, which is also known to be a target of the bovine torovirus HE. A more exciting case of such a switching event occurs in the murine CoVs, with the example of the β-CoV lineage A type binding with two different subtypes of the typical 9-O-Ac-SA (type I) and the exclusive 4-O-Ac-SA (type II) attachment factors. The protein structure data for type II HE also imply the virus switching to binding 4-O acetyl SA from 9-O acetyl SA. Principles of the protein–glycan interaction and PCI stereochemistry potentiate the SA–ligand switch via simple conformational shifts of the lectin and esterase domains. Thus, our understanding of natural adaptation can be specified to how carbohydrate/glycan-recognizing proteins/molecules contribute to virus evolution toward host tropism. Under the current circumstances where reliable antiviral therapeutics or vaccination tools are lacking, several trials are underway to examine viral agents. As expected, structural and non-structural proteins of SARS-CoV-2 are currently being targeted for viral therapeutic designation and development. However, the modern global society needs SARS-CoV-2 preventive and therapeutic drugs for infected patients.
2. Introduction
The recent coronavirus pandemic crisis is due to viral infection of severe acute respiratory syndrome-related coronavirus-2 (y emerged SARS-CoV-2), causing uncontrolled inflammatory conditions in the human lung. Soon after the first transmission emergence of SARS-CoV from animals to humans in China in 2003 [1], a is the cause of the genetically evolved beta-coronavirus genus similar to human viruses was discovered in Chinese horseshoe bats (Rhinolophus sinicus) [2]. To date, pneumonia is epidemiologiobal health cally caused by diverse viruses. For example, adenovirus, influenza virus, Middle East respiratory syndrome virus (MERS-V), parainfluenza virus, respiratory syncytial virus (RSV), SARS-CoV and enteric enveloped CoV can cause pneumonia in human hosts.
Thisis of the World Health Organization (WHO) reported the official terminology of the 2019-Novel Cooronavirus (2019-nCoV) on 13 January 2020, and on February 11th, WHO edited the name of the didisease caused by 2019-nCoV to Coronavirus Disease-2019 2019 (COVID-19). In academia, the International Committee on Taxonomy of Viruses (ICTV) provided official nomenclature to the virus as SARS-CoV-2 due to the similarity between the novel coronavirus and SARS- pandemic. No evidence is yet available for CoV [3]. SARS-CoV-2 is sprnfeading and causing a global health-threatening emergency [4]. Researchers have been ction in a race to develop anti-viral drugs against SARS-CoV-2 even before the WHO declared a worldwide pandemic threatening human lives. Preventive and therapeutic drugs for patients infected with SARS-CoV-2 are yet to be discovered.
The CoVs hosts upon zoonotic diseas enveloped forms can also infect the gastrointestinal track (GIT) outbreak, although most other enteric viruses are naked in morphology [5][6]. the CoVs can also rarely infect neural cells [7]. There is, unfortunatelepidemy, no solid information on how the coronaviruses infect humans and animals with reciprocal infectivity and cause a zoonotic viral outbreak. This is in contrast to inresembles influenza viruses, which are known to selectively utilize use sialic acid (SA) linkages [8]. Currently, only limited information is available on β-CoVs, such as on SARS-CoV-2 and its receptor usage and infectible cell types from different species. Host cell surface s is limited. O-acetylated SAs are recognized by interact with the lectin-like spike glycoproteins of SARS CoV-2 for the first step of attachment to host cells. Infectious virus interaction with the host cell surface is mediated by sialoglycans as the most important phenomenon in eukaryote-parasite co-evolution. O-GlcNAc, a minor glycan source, is mainly found in the nucleus and cytosol (Figure 1). Apart from the general roles of glycans, CoVs recognize host cells and attach to host cell surface molecules to enter tinitial attachment of viruses to enter into the host cells. For example, activity of the heSARS-CoV-2 hemagglutinin-esterase (HE) enzyme relies on the typical carbohydrateacts as the classical glycan-binding lectin and receptor-destroygrading enzyme (RDE) domains. Most β-CoVs targetrecognize 9-O-acetylated -SAs, but certain species have s but switched to recognizing the 4-O-acetyl -SA instead [8][9]. Crystallfographic data for the molecular structure of type II HE provides an explanationm during evolution of CoVs. Type I HE is specific for the swit9-O-Aching mechanism to acquire 4-O acetyl SA binding. This event follows the orthodox ligand–receptor interaction (LRI), lectin–carbohydrate interaction (LCI), lectin–glycan interaction (LGI), lectin–sphingolipid interaction (LSI), protein–glycan interaction (PGI), protein–carbohydrate interaSAs and type II HE is specific for 4-O-Action (PCI), and al-SAso protein–protein interaction (PPI). Recently, 332 protein candidates were suggested to be SARS-CoV-2-human protein interacting protein. The SA-binding shift proceeds through PPIs. Among these, 66 human proteins, as druggable host factors, were further characterized as possible FDA-approvable drugs [10]. If PPI is iquasi-synvolved, however, carbohydrates or glycans may serve as co-receptors or co-determinants. Previous reports suggest that carbohydrates act as receptor determinants in most cases. The general principles of PCI stereochemistry potentiate the SA–ligand switch by way of simple conformational shifts forhronous adaptations of the SA-recognition sites of the lectin and esterase domain. This indicates that our examination of natural adaptation should be dires. The molecular switching of HE acquisition of 4-O-acted to how carbohydrate-btyl binding proteins measure from 9-O-and obscerve carbohydrates, leading to virus evolution toward transitional host tropism.
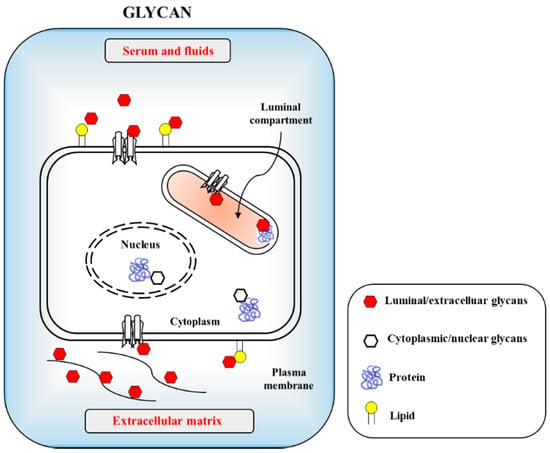
Figure 1.tyl SA binding is Cellular glycans are recognized by infectious agents including viruses and bacteria through used by protein–carbohydrate interaction (PCI) or lectin–carbohydrate interaction (LCI). The carbohydrates are uses as cellular adhesion sites in eukaryotic cells. Host cell surfaced and cytosolic glycans include glycoproteins, glycolipids and proteoglycans with minor glycan species of O-GlcNAc present in nucleus and cytosols.
TheHE gene was transmitted to a β-CoV lineage A progenitor 9-carbon SAs are mainly animal-specific with anionic sugars attached to terminal sugars. SAs exist in two forms, NeuGc and NeuAc. NeuGc is a differentially modified form of the parental Sy horizontal gene transfer from a 9-O-A form, Neu5Ac. SAs are structurally diverse. For example, several modified SA forms are known for their structures including neuraminic acid (NeuC), N-acetyl neuraminic acid (NeuAc), N-glycolyl neuraminic acid (NeuGc), N,O-diacetyl neuraminic acid (occurs in horses), N,O-diacetyl neuraminic acid (occurs in bovines) and N-acetyl O-diacetyl neuraminic acid (occurs in bovines) (Figure 2). Sialyltransferases (STs) biosynthesize different SA linkages. SA linkage diversity occurs at the α2-3, α2-6, α2-8 or α2-9 to the SA or Gal residues (Figure 3). For example, in formation of α2,3 SA or α2,6 SA structures, α2,3-ST and α2,6-ST utilize substrates such as Galβ-1,4-GlcNAc (Figure 4). The most frequent modification of SAs is O-acetyl-SA–specific HEF, as in influenza virus C/D. HE acquisition, and expansion takes place by cross-species transmission over HE evolution. This reflects viral evolutionary adaptation at positions of C4, C7, C8 and C9 of SA (Figure 5).
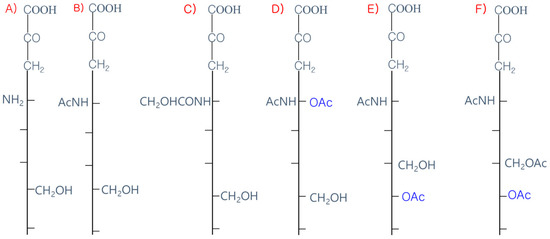
Figure 2. Diverse struto host SA-ctures of sialic acids (SA). (A) Neuraminic tacid; (NeuC); (B) N-acetyl ineuraminic acid (NeuAc); (C) N-g glycolyl neuraminic acid (NeuGc); (D) N, O-diacns. Theretyl nforeuraminic acid (occurs in horse); (E) N, O-dia, CoV HE receptyl neuraminic acid (occurs in bovine); (F) N-aor switching precetyl O-diacetyl neuraminic acid (occurs in bovine).
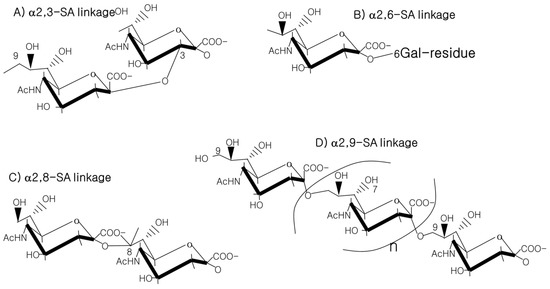
Figure 3. SA linkagees virus of α2-3, α2-6, α2-8 or α2-9 to the SA or Gal residues.
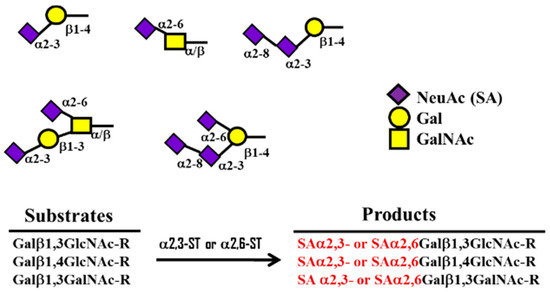
Figure 4. Formavolution of α2,3 ST odr α2,6 SA structures by α 2,3- and 2,6-sialyltransferase (ST) using substrates such as Galβ-1,4-GlcNAc.
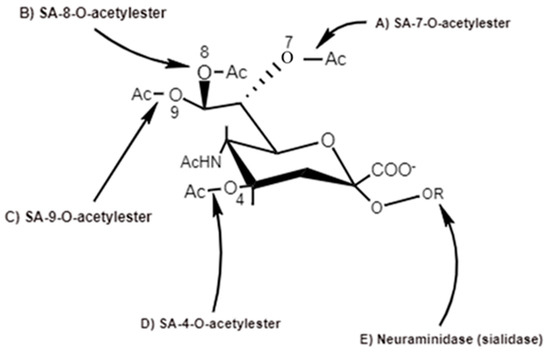
Figure 5. Action sitiven by thes of viral SA-O-acetglylesterases (C4, C7, C8 and C9) specific for 4-O-SA-, 7-O-SA-, 8-O-SA and 9-O-SA and neuraminidases [6].
3. Data, Model, Applications and Influences
3.1. General O-Acetylation of SA
SAs haive various derivatives of more than 50 chemically different structures formed from the basic N-acetylneuraminic acid (Neu5Ac) on the main ring of pyranose and the glycerol side chain. SA are modified by acetyl-, lactyl-, methyl- and sulfo-groups individually or in multiple combinations [11]. Mulsity of the hosts. The PCI or LCI stereochemistry potentiatiple enzymes are involved in the modifications [12]. Hthe SA–ligand swistorically, the first discovered SA was crystallized by Gunner Blix via a hot mild acid extraction of bovine submaxillary mucin in 1936. It consisted of two acetyl groups. Among these, only one acetyl group was attached to nitrogench by a simple conformational shift [13]. Blix isolated a 9-O-acetyl SA of the common SA N-acetylneuraminic acid (Neu5Ac), chemically described as 9-O-aceectyl-N-acetylneuraminic acid (Neu5,9Ac2). Neu5,9Ac2, Neu5Ac and Neu5Gc are naturally occurring SA species in mammals. A common modification is O-acetylation. In fact, O-acetylation of SAs is common in organisms.
and esterase domains. The O-acretyl modification occurs in single positions of C-4, C-7, C-8 and C-9 of SA as well as in combined C-positions to yield Neu4,5Ac2, Neu5,9Ac2 and Neu5,7,9Ac3 SAs. Neu5Ac9NAc is a chemical and biologic mimic of Neu5,9Ac2 in the SA-glycans. The C-7 and/or C-9 O-acetylations are catalyzed by the SA O-acetyltransferase enzyme, Cas1 domain containing 1 (CasD1) (Figure 6). CasD1 catalyzes the addition of acetyl groups to the SA C-7 at the late Golgi apparatus compartmentfore, examination of new emerging viruses can lead to better understanding of virus evolution [14]. Thereafter, owan enzyme termed “migrase” transfers the addrd transitional acetyl-group from C-7 to C-9, although this enzyme has not been identified [12]host tropism. CasD1A uses acetyl-coenzyme A as a donor substrate and CMP-Neu5Ac as an acceptor substrate, but with weak activity on CMP-Neu5Gc.
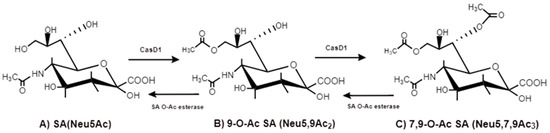
Figure 6.clear example of HE gene CasD1, SA O-acetyltransferase, transfers acetyl groups to C7 position of SA (Neu5Ac), from which it migrates to the C9 position (Neu5,9Ac2). The additional acetyl group is added to C7 of SA (Neu5,7,9 is found in the BCoV HE, which prefers 7,9-di-O-Ac3) by the same CasD1. The -SA O-acetylesterase cleaves of the acetyl groups.
Bs, whiologically, the SA O-acetylation event confers merits to hosts such as protection from pathogenic invasion and maintenance of systemic self-homeostasis. The O-acetylation event of SAs protects the SA-containing glycans from neuraminidase (NA)/sialidase action, because O-acetyl-groups inhibit microbial NA activity. The chemical structure of the O-acetyl group is quite unstable and susceptible to esterase enzymes. Sialic acid cleavageh is also known to be a target of the bovine torovirus HE. A more exciting case of such a switching event occurs in the murine CoVs, with the example of the di-acetylated Neu5,7,9Ac3 by bacterial NAs decreases two-fold, when compared to mono-O-β-CoV lineacgetylated Neu5Ac. The O-acetylated glycan modification invites interaction with viruses, antibodies and mammalian lectins [15]. Therefore, A type binding withe SA O-acetylation modification confers specific functions to organisms.
3.2. Evolutionary Acquisition of C4-O-Acetyl and C9-O-Acetyl SA Recognition by HE Enzymes
3.2.1. C4-O-Acetyl Modification
For examtwo different subtyple, in the horse, C4-O-aces of the tyl modpification of Neu5cal 9-O-Ac (-SA) occupies more than 50% of the total SA content. The C (type I) and the exclusive 4-O-acetylated Neu5Ac, Neu4,5Ac2, inhibits the influenza A2 virus HA. De-acetylation reagents such as NaOH or NaIO4-SA (type II) treatment completely hemagglutinate Neu4,5Ac2 by elimination of the C4-O-tachment facetyl group [16]ors. The C4-O-acetyl Neu5Ac species are found in various sources such as equine erythrocyte GM3, starfish A. rubens anotein structure d fish [17][18][19][20][21]. C4-O-acetylated Neu5Ac facilitates the initial attachment of viruses to target cells. Like the influenza C virus, infectious salmon anemia virus (ISAV), a member of the Orthomyxoviridae family, contains HE and HEF proteins to mediate a for type II HE also imply the virus entry and exit. C4-O-Aswitc Neu5Ac is the major receptor determinant of ISAV in receptoring to binding and destruction [21], whi 4-O acetyle the influenza C virus recognizes CSA from 9-O-AcO Neu5Ac. The acetylesterase RDE of ISAV cleaves C4-O-Ac via 4-SA-O-acetyleste SA. Prase with a short turnover time, whereas C9-O-Anc Neu5Ac is cleaved by 9-SA-O-acetyleplesterase with a long turnover time [17].
Tof the prosition of SA O-acetylation is linked to functions including substrate differentiation of enzymes such as NAs and esterase by C4 O-Ac. Previous development of O-Ac site-selective NA inhibitors were based on the conceptual consideration of different O-Ac positions. The O-Ac of SAs is site-specific, as C4 of Neu5Ac is considered to be a potential position for modification. Historically, inhibitors of influenza A and B viruses-sialidases were designed by Von-Itzstein in 1993 [22]tein–glycan interaction and PCI stereochemistry potentiate the SA–ligand switch via simple conformational shifts of the lectin and esterase domains. The Acus, group-based C4 substitution interacts with amino acid Glu-119 present in the active site of sialidase. Guanidine-attached C4 of C2–C3 unsaturated SA (Neu5Ac2en) inhibits activity of sialidases isolated from influenza A virus (Singapore/1/57) and B virus (Victoria/102/85). The same scenario was applied for sialidase inhibition of the human parainfluenza virus type 3, which has HN and fusion our understanding of natural adaptation can be specified to how carbohydrate/glycan-recognizing proteins [23]. The C4 /mof Nleu5Ac2en was substituted by alkyl groups such as the O-ethyl group. For example, Zanamivir has a substitution with a 4-guanidino group with an IC50 of 25 μM. Thus, sialidase inhibition is important for C4 modification of Neu5Ac2en. Later, oseltamivir with the tradename Tamiflu (Basel, Switzerland) and zanamivir with the tradename Relenza (London, UK) were establishedcules contribute to virus evolution toward host tropism. Under the current circumstances where reliable [24]. These drugs exhibantit some adverse side effects that restrict clinical use.
3.2.2. SA C9-O-Acetyl Modification
Tiral the SA 9-O-racpetylation in hosts allows hosts to evade influenza A virus hemagglutinin (HA) recognition and some lectins of factor H (FH), CD22/Siglec-2 and sialoadhesin/Siglec-1. Instead, the influenza C virus HA recognizes the hosts. β-elimination and permethylation eliminate the 9-O-acetyl gutics or vaccination tools are lacking, several troup from SAs. Chemical modification of the C-9 position of Neu5,9Ac2 generates a 9-N-acetyl analog, 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc), a mimic of Neu5,9Ac2 with influenza C virus-binding capacity, which is not cleaved by the HE [25]als are underway to examine viral agents. SA O-acetylesterase regulates the presence of 7,9-O-Ac and 9-O-Ac. SA O-aces expected, structyluration and deacetylation are involved in development, cancer and immunology. SA O-acetylation alters host lectin bindings such as siglecsl and non-structural [12]. The presence of 9-O-Ac can also reduce the activity of NAs [26]. ins of SARSA m-Codifications regulate pathogen binding or pathogen NAs. Influenza A/B/C/D viruses use SA as their entry receptors. Influenza A and B subtypes bind to SAs via HA and NA to allow endocytosis of the virus and fusion of the viral envelope with endosomes. In contrast, influenza C and D subtypes bear only one coated glycoprotein, termed the HE fusion protein (HEF). The HEF acts as the HA and NA. HEF recognizes 9-O-V-2 are currently being targeted for viral therapeutic designation and development. However, the modern globacetyl SA for entry into cells, while the esterase domain removes 9-O-acsociety needs SARS-CoV-2 preventivetyl-groups and liberates the virus from mucus and mis-assembled virus aggregates after budding. The 9-O-Atherapeutic drugs for infec on ctells prevents the NA activity and HA binding of the influenza A type virus [27]d patients.





3. Data, Model, Applications and Influences
3.1. General O-Acetylation of SA
SAs have various derivatives of more than 50 chemically different structures formed from the basic N-acetylneuraminic acid (Neu5Ac) on the main ring of pyranose and the glycerol side chain. SA are modified by acetyl-, lactyl-, methyl- and sulfo-groups individually or in multiple combinations [28]. Multiple enzymes are involved in the modifications [29]. Historically, the first discovered SA was crystallized by Gunner Blix via a hot mild acid extraction of bovine submaxillary mucin in 1936. It consisted of two acetyl groups. Among these, only one acetyl group was attached to nitrogen [30]. Blix isolated a 9-O-acetyl SA of the common SA N-acetylneuraminic acid (Neu5Ac), chemically described as 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2). Neu5,9Ac2, Neu5Ac and Neu5Gc are naturally occurring SA species in mammals. A common modification is O-acetylation. In fact, O-acetylation of SAs is common in organisms.
The O-acetyl modification occurs in single positions of C-4, C-7, C-8 and C-9 of SA as well as in combined C-positions to yield Neu4,5Ac2, Neu5,9Ac2 and Neu5,7,9Ac3 SAs. Neu5Ac9NAc is a chemical and biologic mimic of Neu5,9Ac2 in the SA-glycans. The C-7 and/or C-9 O-acetylations are catalyzed by the SA O-acetyltransferase enzyme, Cas1 domain containing 1 (CasD1) (Figure 6). CasD1 catalyzes the addition of acetyl groups to the SA C-7 at the late Golgi apparatus compartment [31]. Thereafter, an enzyme termed “migrase” transfers the additional acetyl-group from C-7 to C-9, although this enzyme has not been identified [29]. CasD1 uses acetyl-coenzyme A as a donor substrate and CMP-Neu5Ac as an acceptor substrate, but with weak activity on CMP-Neu5Gc.
 Figure 6. CasD1, SA O-acetyltransferase, transfers acetyl groups to C7 position of SA (Neu5Ac), from which it migrates to the C9 position (Neu5,9Ac2). The additional acetyl group is added to C7 of SA (Neu5,7,9Ac3) by the same CasD1. The SA O-acetylesterase cleaves of the acetyl groups.
Biologically, the SA O-acetylation event confers merits to hosts such as protection from pathogenic invasion and maintenance of systemic self-homeostasis. The O-acetylation event of SAs protects the SA-containing glycans from neuraminidase (NA)/sialidase action, because O-acetyl-groups inhibit microbial NA activity. The chemical structure of the O-acetyl group is quite unstable and susceptible to esterase enzymes. Sialic acid cleavage of the di-acetylated Neu5,7,9Ac3 by bacterial NAs decreases two-fold, when compared to mono-O-acetylated Neu5Ac. The O-acetylated glycan modification invites interaction with viruses, antibodies and mammalian lectins [32]. Therefore, the SA O-acetylation modification confers specific functions to organisms.
Figure 6. CasD1, SA O-acetyltransferase, transfers acetyl groups to C7 position of SA (Neu5Ac), from which it migrates to the C9 position (Neu5,9Ac2). The additional acetyl group is added to C7 of SA (Neu5,7,9Ac3) by the same CasD1. The SA O-acetylesterase cleaves of the acetyl groups.
Biologically, the SA O-acetylation event confers merits to hosts such as protection from pathogenic invasion and maintenance of systemic self-homeostasis. The O-acetylation event of SAs protects the SA-containing glycans from neuraminidase (NA)/sialidase action, because O-acetyl-groups inhibit microbial NA activity. The chemical structure of the O-acetyl group is quite unstable and susceptible to esterase enzymes. Sialic acid cleavage of the di-acetylated Neu5,7,9Ac3 by bacterial NAs decreases two-fold, when compared to mono-O-acetylated Neu5Ac. The O-acetylated glycan modification invites interaction with viruses, antibodies and mammalian lectins [32]. Therefore, the SA O-acetylation modification confers specific functions to organisms.
3.2. Evolutionary Acquisition of C4-O-Acetyl and C9-O-Acetyl SA Recognition by HE Enzymes
3.2.1. C4-O-Acetyl Modification
For example, in the horse, C4-O-acetyl modification of Neu5Ac (SA) occupies more than 50% of the total SA content. The C4-O-acetylated Neu5Ac, Neu4,5Ac2, inhibits the influenza A2 virus HA. De-acetylation reagents such as NaOH or NaIO4 treatment completely hemagglutinate Neu4,5Ac2 by elimination of the C4-O-acetyl group [33]. The C4-O-acetyl Neu5Ac species are found in various sources such as equine erythrocyte GM3, starfish A. rubens and fish [34,35,36,37,38]. C4-O-acetylated Neu5Ac facilitates the initial attachment of viruses to target cells. Like the influenza C virus, infectious salmon anemia virus (ISAV), a member of the Orthomyxoviridae family, contains HE and HEF proteins to mediate virus entry and exit. C4-O-Ac Neu5Ac is the major receptor determinant of ISAV in receptor binding and destruction [38], while the influenza C virus recognizes C9-O-Ac Neu5Ac. The acetylesterase RDE of ISAV cleaves C4-O-Ac via 4-SA-O-acetylesterase with a short turnover time, whereas C9-O-Ac Neu5Ac is cleaved by 9-SA-O-acetylesterase with a long turnover time [34].
The position of SA O-acetylation is linked to functions including substrate differentiation of enzymes such as NAs and esterase by C4 O-Ac. Previous development of O-Ac site-selective NA inhibitors were based on the conceptual consideration of different O-Ac positions. The O-Ac of SAs is site-specific, as C4 of Neu5Ac is considered to be a potential position for modification. Historically, inhibitors of influenza A and B viruses-sialidases were designed by Von-Itzstein in 1993 [39]. The Ac group-based C4 substitution interacts with amino acid Glu-119 present in the active site of sialidase. Guanidine-attached C4 of C2–C3 unsaturated SA (Neu5Ac2en) inhibits activity of sialidases isolated from influenza A virus (Singapore/1/57) and B virus (Victoria/102/85). The same scenario was applied for sialidase inhibition of the human parainfluenza virus type 3, which has HN and fusion proteins [40]. The C4 of Neu5Ac2en was substituted by alkyl groups such as the O-ethyl group. For example, Zanamivir has a substitution with a 4-guanidino group with an IC50 of 25 μM. Thus, sialidase inhibition is important for C4 modification of Neu5Ac2en. Later, oseltamivir with the tradename Tamiflu (Basel, Switzerland) and zanamivir with the tradename Relenza (London, UK) were established [41]. These drugs exhibit some adverse side effects that restrict clinical use.
3.2.2. SA C9-O-Acetyl Modification
The SA 9-O-acetylation in hosts allows hosts to evade influenza A virus hemagglutinin (HA) recognition and some lectins of factor H (FH), CD22/Siglec-2 and sialoadhesin/Siglec-1. Instead, the influenza C virus HA recognizes the hosts. β-elimination and permethylation eliminate the 9-O-acetyl group from SAs. Chemical modification of the C-9 position of Neu5,9Ac2 generates a 9-N-acetyl analog, 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc), a mimic of Neu5,9Ac2 with influenza C virus-binding capacity, which is not cleaved by the HE [42]. SA O-acetylesterase regulates the presence of 7,9-O-Ac and 9-O-Ac. SA O-acetylation and deacetylation are involved in development, cancer and immunology. SA O-acetylation alters host lectin bindings such as siglecs [29]. The presence of 9-O-Ac can also reduce the activity of NAs [43]. SA modifications regulate pathogen binding or pathogen NAs. Influenza A/B/C/D viruses use SA as their entry receptors. Influenza A and B subtypes bind to SAs via HA and NA to allow endocytosis of the virus and fusion of the viral envelope with endosomes. In contrast, influenza C and D subtypes bear only one coated glycoprotein, termed the HE fusion protein (HEF). The HEF acts as the HA and NA. HEF recognizes 9-O-acetyl SA for entry into cells, while the esterase domain removes 9-O-acetyl-groups and liberates the virus from mucus and mis-assembled virus aggregates after budding. The 9-O-Ac on cells prevents the NA activity and HA binding of the influenza A type virus [44].
References
- Varki, A.; Schauer, R. Sialic Acids: Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Mandal, C.; Schwartz-Albiez, R.; Vlasak, R. Functions and Biosynthesis of O-Acetylated Sialic Acids. Topics Curr. Chem. 2012, 366, 1–30. [Google Scholar] [CrossRef]
- Lundblad, A. Gunnar Blix and his discovery of sialic acids. Fascinating molecules in glycobiology. Upsala J. Med Sci. 2015, 120, 104–112. [Google Scholar] [CrossRef]
- Baumann, A.M.T.; Bakkers, M.J.G.; Buettner, F.F.R.; Hartmann, M.; Grove, M.; Langereis, M.A.; de Groot, R.J.; Mühlenhoff, M. 9-O-Acetylation of sialic acids is catalyzed by CASD1 via a covalent acetyl-enzyme intermediate. Nat. Commun. 2015, 6, 7673. [Google Scholar] [CrossRef]
- Khedri, Z.; Xiao, A.; Yu, H.; Landig, C.S.; Li, W.; Diaz, S.; Wasik, B.R.; Parrish, C.R.; Wang, L.-P.; Varki, A.; et al. A Chemical Biology Solution to Problems with Studying Biologically Important but Unstable 9-O-Acetyl Sialic Acids. ACS Chem. Boil. 2016, 12, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Srinivasan, G.V.; Wipfler, D.; Kniep, B.; Schwartz-Albiez, R. O-Acetylated Sialic Acids and Their Role in Immune Defense. Adv. Exp. Med. Biol. 2011, 705, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S. Post-Glycosylation Modification of Sialic Acid, and Its Role in Virus Pathogenesis. Vaccines 2019, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Yachida, Y.; Tsuchihashi, K.; Gasa, S. Novel di-O-acetylated GM3s from equine erythrocytes, one containing 4,9-di-O-acetyl-N-glycolylneuraminic acid and another containing 4-O-acetyl-N-glycolylneuraminic acid and 6-O-acetyl-D-galactose. Carbohydr. Res. 1997, 298, 201–212. [Google Scholar] [CrossRef]
- Zanetta, J.-P.; Srinivasan, V.; Schauer, R. Analysis of monosaccharides, fatty constituents, and rare O-acetylated sialic acids from gonads of the starfish Asterias rubens. Biochimie 2006, 88, 171–178. [Google Scholar] [CrossRef]
- Aamelfot, M.; Dale, O.B.; Weli, S.C.; Koppang, E.O.; Falk, K. The in-situ distribution of glycoprotein-bound 4-O-Acetylated sialic acids in vertebrates. Glycoconj. J. 2014, 31, 327–335. [Google Scholar] [CrossRef]
- Hellebø, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious Salmon Anemia Virus Specifically Binds to and Hydrolyzes 4-O-Acetylated Sialic Acids. J. Virol. 2004, 78, 3055–3062. [Google Scholar] [CrossRef]
- von Itzstein, M.; Wu, W.-Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; van Phan, T.; Smythe, M.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar] [CrossRef]
- Tindal, D.J.; Dyason, J.C.; Thomson, R.J.; Suzuki, T.; Ueyama, H.; Kuwahara, Y.; Maki, N.; Suzuki, Y.; von Itzstein, M. Synthesis and evaluation of 4-O-alkylated 2-deoxy-2,3-didehydro-N-acetylneuraminic acid derivatives as inhibitors of human parainfluenza virus type-3 sialidase activity. Bioorg. Med. Chem. Lett. 2007, 17, 1655–1658. [Google Scholar] [CrossRef]
- Yu, K.; Luo, C.; Qin, G.; Xu, Z.; Li, N.; Liu, H.; Shen, X.; Ma, J.; Wang, Q.; Yang, C.; et al. Why are Oseltamivir and Zanamivir effective against the newly emerged influenza A virus (A/H1N1)? Cell Res. 2009, 19, 1221–1224. [Google Scholar] [CrossRef]
- Herrler, G.; Gross, H.J.; Imhof, A.; Brossmer, R.; Milks, G.; Paulson, J.C. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J. Boil. Chem. 1992, 267, 12501–12505. [Google Scholar]
- Hunter, C.D.; Khanna, N.; Richards, M.; Darestani, R.R.; Zou, C.; Klassen, J.S.; Cairo, C.W. Human Neuraminidase Isoenzymes Show Variable Activities for 9-O-Acetyl-sialoside Substrates. ACS Chem. Boil. 2018, 13, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Barnard, K.N.; Wasik, B.R.; la Clair, J.R.; Buchholz, D.W.; Weichert, W.S.; Alford-Lawrence, B.K.; Aguilar, H.C.; Parrish, C.R. Expression of 9-O- and 7,9-O-Acetyl Modified Sialic Acid in Cells and Their Effects on Influenza Viruses. mBio 2019, 10, 02490-19. [Google Scholar] [CrossRef] [PubMed]
3.1. General O-Acetylation of SA

3.2. Evolutionary Acquisition of C4-O-Acetyl and C9-O-Acetyl SA Recognition by HE Enzymes
3.2.1. C4-O-Acetyl Modification
3.2.2. SA C9-O-Acetyl Modification
References
- Varki, A.; Schauer, R. Sialic Acids: Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Mandal, C.; Schwartz-Albiez, R.; Vlasak, R. Functions and Biosynthesis of O-Acetylated Sialic Acids. Topics Curr. Chem. 2012, 366, 1–30. [Google Scholar] [CrossRef]
- Lundblad, A. Gunnar Blix and his discovery of sialic acids. Fascinating molecules in glycobiology. Upsala J. Med Sci. 2015, 120, 104–112. [Google Scholar] [CrossRef]
- Baumann, A.M.T.; Bakkers, M.J.G.; Buettner, F.F.R.; Hartmann, M.; Grove, M.; Langereis, M.A.; de Groot, R.J.; Mühlenhoff, M. 9-O-Acetylation of sialic acids is catalyzed by CASD1 via a covalent acetyl-enzyme intermediate. Nat. Commun. 2015, 6, 7673. [Google Scholar] [CrossRef]
- Khedri, Z.; Xiao, A.; Yu, H.; Landig, C.S.; Li, W.; Diaz, S.; Wasik, B.R.; Parrish, C.R.; Wang, L.-P.; Varki, A.; et al. A Chemical Biology Solution to Problems with Studying Biologically Important but Unstable 9-O-Acetyl Sialic Acids. ACS Chem. Boil. 2016, 12, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Srinivasan, G.V.; Wipfler, D.; Kniep, B.; Schwartz-Albiez, R. O-Acetylated Sialic Acids and Their Role in Immune Defense. Adv. Exp. Med. Biol. 2011, 705, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S. Post-Glycosylation Modification of Sialic Acid, and Its Role in Virus Pathogenesis. Vaccines 2019, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Yachida, Y.; Tsuchihashi, K.; Gasa, S. Novel di-O-acetylated GM3s from equine erythrocytes, one containing 4,9-di-O-acetyl-N-glycolylneuraminic acid and another containing 4-O-acetyl-N-glycolylneuraminic acid and 6-O-acetyl-D-galactose. Carbohydr. Res. 1997, 298, 201–212. [Google Scholar] [CrossRef]
- Zanetta, J.-P.; Srinivasan, V.; Schauer, R. Analysis of monosaccharides, fatty constituents, and rare O-acetylated sialic acids from gonads of the starfish Asterias rubens. Biochimie 2006, 88, 171–178. [Google Scholar] [CrossRef]
- Aamelfot, M.; Dale, O.B.; Weli, S.C.; Koppang, E.O.; Falk, K. The in-situ distribution of glycoprotein-bound 4-O-Acetylated sialic acids in vertebrates. Glycoconj. J. 2014, 31, 327–335. [Google Scholar] [CrossRef]
- Hellebø, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious Salmon Anemia Virus Specifically Binds to and Hydrolyzes 4-O-Acetylated Sialic Acids. J. Virol. 2004, 78, 3055–3062. [Google Scholar] [CrossRef]
- von Itzstein, M.; Wu, W.-Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; van Phan, T.; Smythe, M.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar] [CrossRef]
- Tindal, D.J.; Dyason, J.C.; Thomson, R.J.; Suzuki, T.; Ueyama, H.; Kuwahara, Y.; Maki, N.; Suzuki, Y.; von Itzstein, M. Synthesis and evaluation of 4-O-alkylated 2-deoxy-2,3-didehydro-N-acetylneuraminic acid derivatives as inhibitors of human parainfluenza virus type-3 sialidase activity. Bioorg. Med. Chem. Lett. 2007, 17, 1655–1658. [Google Scholar] [CrossRef]
- Yu, K.; Luo, C.; Qin, G.; Xu, Z.; Li, N.; Liu, H.; Shen, X.; Ma, J.; Wang, Q.; Yang, C.; et al. Why are Oseltamivir and Zanamivir effective against the newly emerged influenza A virus (A/H1N1)? Cell Res. 2009, 19, 1221–1224. [Google Scholar] [CrossRef]
- Herrler, G.; Gross, H.J.; Imhof, A.; Brossmer, R.; Milks, G.; Paulson, J.C. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J. Boil. Chem. 1992, 267, 12501–12505. [Google Scholar]
- Hunter, C.D.; Khanna, N.; Richards, M.; Darestani, R.R.; Zou, C.; Klassen, J.S.; Cairo, C.W. Human Neuraminidase Isoenzymes Show Variable Activities for 9-O-Acetyl-sialoside Substrates. ACS Chem. Boil. 2018, 13, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Barnard, K.N.; Wasik, B.R.; la Clair, J.R.; Buchholz, D.W.; Weichert, W.S.; Alford-Lawrence, B.K.; Aguilar, H.C.; Parrish, C.R. Expression of 9-O- and 7,9-O-Acetyl Modified Sialic Acid in Cells and Their Effects on Influenza Viruses. mBio 2019, 10, 02490-19. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Schauer, R. Sialic Acids: Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Mandal, C.; Schwartz-Albiez, R.; Vlasak, R. Functions and Biosynthesis of O-Acetylated Sialic Acids. Topics Curr. Chem. 2012, 366, 1–30. [Google Scholar] [CrossRef]
- Lundblad, A. Gunnar Blix and his discovery of sialic acids. Fascinating molecules in glycobiology. Upsala J. Med Sci. 2015, 120, 104–112. [Google Scholar] [CrossRef]
- Baumann, A.M.T.; Bakkers, M.J.G.; Buettner, F.F.R.; Hartmann, M.; Grove, M.; Langereis, M.A.; de Groot, R.J.; Mühlenhoff, M. 9-O-Acetylation of sialic acids is catalyzed by CASD1 via a covalent acetyl-enzyme intermediate. Nat. Commun. 2015, 6, 7673. [Google Scholar] [CrossRef]
- Khedri, Z.; Xiao, A.; Yu, H.; Landig, C.S.; Li, W.; Diaz, S.; Wasik, B.R.; Parrish, C.R.; Wang, L.-P.; Varki, A.; et al. A Chemical Biology Solution to Problems with Studying Biologically Important but Unstable 9-O-Acetyl Sialic Acids. ACS Chem. Boil. 2016, 12, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Srinivasan, G.V.; Wipfler, D.; Kniep, B.; Schwartz-Albiez, R. O-Acetylated Sialic Acids and Their Role in Immune Defense. Adv. Exp. Med. Biol. 2011, 705, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S. Post-Glycosylation Modification of Sialic Acid, and Its Role in Virus Pathogenesis. Vaccines 2019, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Yachida, Y.; Tsuchihashi, K.; Gasa, S. Novel di-O-acetylated GM3s from equine erythrocytes, one containing 4,9-di-O-acetyl-N-glycolylneuraminic acid and another containing 4-O-acetyl-N-glycolylneuraminic acid and 6-O-acetyl-D-galactose. Carbohydr. Res. 1997, 298, 201–212. [Google Scholar] [CrossRef]
- Zanetta, J.-P.; Srinivasan, V.; Schauer, R. Analysis of monosaccharides, fatty constituents, and rare O-acetylated sialic acids from gonads of the starfish Asterias rubens. Biochimie 2006, 88, 171–178. [Google Scholar] [CrossRef]
- Aamelfot, M.; Dale, O.B.; Weli, S.C.; Koppang, E.O.; Falk, K. The in-situ distribution of glycoprotein-bound 4-O-Acetylated sialic acids in vertebrates. Glycoconj. J. 2014, 31, 327–335. [Google Scholar] [CrossRef]
- Hellebø, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious Salmon Anemia Virus Specifically Binds to and Hydrolyzes 4-O-Acetylated Sialic Acids. J. Virol. 2004, 78, 3055–3062. [Google Scholar] [CrossRef]
- von Itzstein, M.; Wu, W.-Y.; Kok, G.B.; Pegg, M.S.; Dyason, J.C.; Jin, B.; van Phan, T.; Smythe, M.; White, H.F.; Oliver, S.W.; et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [Google Scholar] [CrossRef]
- Tindal, D.J.; Dyason, J.C.; Thomson, R.J.; Suzuki, T.; Ueyama, H.; Kuwahara, Y.; Maki, N.; Suzuki, Y.; von Itzstein, M. Synthesis and evaluation of 4-O-alkylated 2-deoxy-2,3-didehydro-N-acetylneuraminic acid derivatives as inhibitors of human parainfluenza virus type-3 sialidase activity. Bioorg. Med. Chem. Lett. 2007, 17, 1655–1658. [Google Scholar] [CrossRef]
- Yu, K.; Luo, C.; Qin, G.; Xu, Z.; Li, N.; Liu, H.; Shen, X.; Ma, J.; Wang, Q.; Yang, C.; et al. Why are Oseltamivir and Zanamivir effective against the newly emerged influenza A virus (A/H1N1)? Cell Res. 2009, 19, 1221–1224. [Google Scholar] [CrossRef]
- Herrler, G.; Gross, H.J.; Imhof, A.; Brossmer, R.; Milks, G.; Paulson, J.C. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J. Boil. Chem. 1992, 267, 12501–12505. [Google Scholar]
- Hunter, C.D.; Khanna, N.; Richards, M.; Darestani, R.R.; Zou, C.; Klassen, J.S.; Cairo, C.W. Human Neuraminidase Isoenzymes Show Variable Activities for 9-O-Acetyl-sialoside Substrates. ACS Chem. Boil. 2018, 13, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Barnard, K.N.; Wasik, B.R.; la Clair, J.R.; Buchholz, D.W.; Weichert, W.S.; Alford-Lawrence, B.K.; Aguilar, H.C.; Parrish, C.R. Expression of 9-O- and 7,9-O-Acetyl Modified Sialic Acid in Cells and Their Effects on Influenza Viruses. mBio 2019, 10, 02490-19. [Google Scholar] [CrossRef] [PubMed]
