LIN28 inhibits let-7 miRNA maturation which prevents cell differentiation and promotes proliferation. We hypothesized that the LIN28-let-7 axis regulates proliferation-associated genes in sheep trophectoderm in vivo. Day 9-hatched sheep blastocysts were incubated with lentiviral particles to deliver shRNA targeting LIN28 specifically to trophectoderm cells. At day 16, conceptus elongation was significantly reduced in LIN28A and LIN28B knockdowns. Let-7 miRNAs were significantly increased and IGF2BP1-3, HMGA1, ARID3B, and c-MYC were decreased in trophectoderm from knockdown conceptuses. Ovine trophoblast (OTR) cells derived from day 16 trophectoderm are a useful tool for in vitro experiments. Surprisingly, LIN28 was significantly reduced and let-7 miRNAs increased after only a few passages of OTR cells, suggesting these passaged cells represent a more differentiated phenotype. To create an OTR cell line more similar to day 16 trophectoderm we overexpressed LIN28A and LIN28B, which significantly decreased let-7 miRNAs and increased IGF2BP1-3, HMGA1, ARID3B, and c-MYC compared to control. This is the first study showing the role of the LIN28-let-7 axis in trophoblast proliferation and conceptus elongation in vivo. These results suggest that reduced LIN28 during early placental development can lead to reduced trophoblast proliferation and sheep conceptus elongation at a critical period for successful establishment of pregnancy.
Embryonic and fetal mortality results in huge economic losses in sheep and cattle industry. Pregnancy loss between fertilization and term birth is 40-55% in cattle and 30-40% in sheep. In domestic ruminants, before attachment to uterus, the hatched blastocyst elongates due to rapid proliferation of trophoblast cells and adopts a filamentous shape comprised of mainly extraembryonic trophoblast cells. Interestingly, majority of pregnancy loss in ruminants occurs during first 2-3 weeks of pregnancy, which is the window of conceptus elongation. Conceptus elongation is critical for implantation, placentation, establishment of pregnancy and foetal development. Reduced conceptus elongation can result into pregnancy loss or intrauterine growth restriction. LIN28 is an RNA binding protein which repress let-7 miRNAs biogenesis. Let-7 miRNAs are markers of cell differentiation and high let-7 levels reduce cell proliferation. Incubating day 9 hatched sheep blastocysts with shRNA-expressing lentiviral particles leads infection of only trophectoderm and spares all other cell types, generating a trophectoderm-specific gene modification. LIN28 knockdown in trophectoderm led to reduced conceptus elongation at 16 dGA compared to control, suggesting reduced proliferation of trophoblast cells. Let-7 miRNAs were significantly increased and proliferation-associated proteins IGF2BP1-3, HMGA1, ARID3B and c-MYC were significantly decreased in trophectoderm from knockdown conceptuses compared to control. This suggests that the LIN28-let-7 axis regulates proliferation of sheep trophoblast cells by targeting proliferation-associated genes.
- Blastocyst
- Conceptus Elongation
- Proliferation
1.LIN28 Knockdown Introducin Trophectoderm Resulted in Reduced Proliferation of Trophoblast Cells and Lower Expression of Proliferation-Associated Genes
1.1. LIN28 Knockdown in Trophectoderm Resulted in Reduced Proliferation of Trophoblast Cells and Lower Expression of Proliferation-Associated Genes
Day 9-hatched blastocysts were infected with lentiviral particles expressing shRNA to knockdown LIN28A (AKD) or LIN28B (BKD), or scramble control shRNA (SC). The blastocysts treated with lentiviral particles were surgically transferred to synchronized ewes at day 9 of estrus. The conceptuses were collected at day 16 of gestation and trophectoderm (TE) was separated from embryo. LIN28A and LIN28B mRNAs and proteins were quantified by real-time PCR and Western blot. LIN28A mRNA and protein was significantly reduced in AKD TE while LIN28B mRNA and protein was significantly reduced in BKD TE compared to SC (Figure 1A,B). As expected, due to reduced LIN28,
Day 9-hatched blastocysts were infected with lentiviral particles expressing shRNA to knockdown LIN28A (AKD) or LIN28B (BKD), or scramble control shRNA (SC). The blastocysts treated with lentiviral particles were surgically transferred to synchronized ewes at day 9 of estrus. The conceptuses were collected at day 16 of gestation and trophectoderm (TE) was separated from embryo. LIN28A and LIN28B mRNAs and proteins were quantified by real-time PCR and Western blot. LIN28A mRNA and protein was significantly reduced in AKD TE while LIN28B mRNA and protein was significantly reduced in BKD TE compared to SC (Figure 1A,B). As expected, due to reduced LIN28,let-7
miRNAs (let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i
) were significantly higher in AKD and BKD TE compared to SC, and there was no significant change inlet-7
miRNAs between AKD and BKD TE (Figure 1C). These results suggest that reduced LIN28A or LIN28B led to significant increase inlet-7 miRNAs.
miRNAs.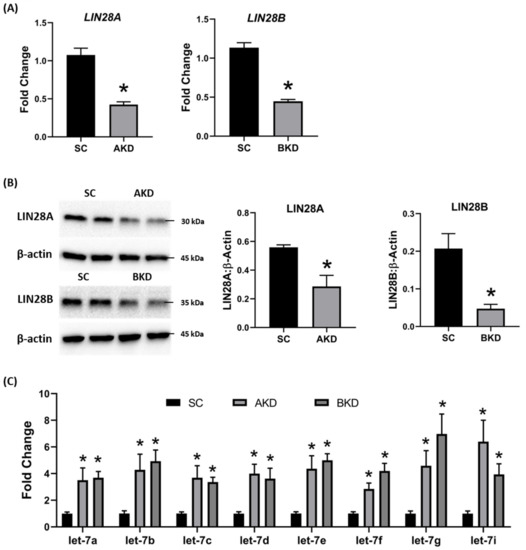
Figure 1.
let-7
A
LIN28A
LIN28B
n
n
n
B
C
Let-7
p < 0.05 vs. SC.
To determine the effect of LIN28 knockdown on conceptus elongation, we measured the length of TE. Knockdown of LIN28A or LIN28B resulted in a significant reduction in day 16 TE length compared to SC (Figure 2A,B). There was no significant difference in elongation of AKD vs. BKD TE. This data suggest knockdown of either LIN28A or LIN28B in vivo resulted in reduced proliferation of trophoblast cells.
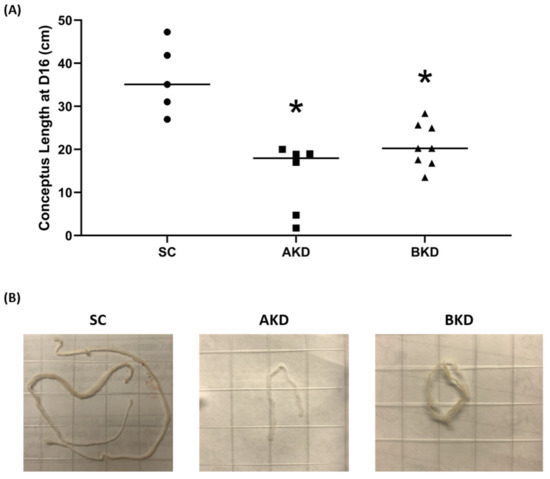
Figure 2.
A
n
n
n
B
p < 0.05 vs. SC.
Due to the potential for reduced proliferation of trophoblast cells in the AKD and BKD conceptuses, we measured the mRNA and protein levels of
let-7
let-7 miRNAs in AKD and BKD TE led to a significant reduction in expression of IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B, and c-MYC, and the reduced proliferation of trophoblast cells in AKD and BKD TE is due to significantly reduced expression of these proliferation-associated genes.
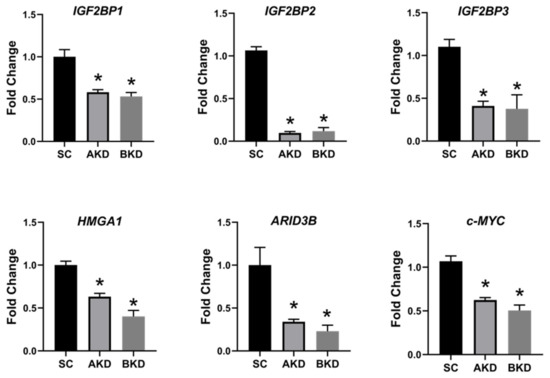
Figure 3.
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC
n
p < 0.05 vs. SC.
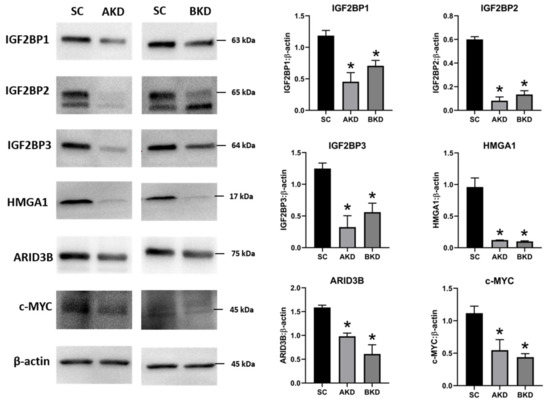
Figure 4.
n
p < 0.05 vs. SC.
2. Ovine Trophoblast Cells Generated from Day 16 Trophectoderm Had a Significant Reduction in LIN28
To further investigate the regulation of ovine trophoblast cell proliferation by LIN28 in vitro, we used day 16 TE to generate ovine trophoblast cells. The day 16 TE was minced and plated in collagen-coated plates and was passaged to obtain a cell line. The cells used for further experiments were collected at passage 4–6, so we called these cells non-immortalized ovine trophoblast (OTR) cells. Interestingly, real-time PCR data showed that OTR cells had a significant reduction in
LIN28A
LIN28B
let-7
let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i
let-7 miRNAs in OTR cells after only 4–6 passages suggest that these cells differentiated to a different phenotype compared to trophoblast cells in day 16 TE.
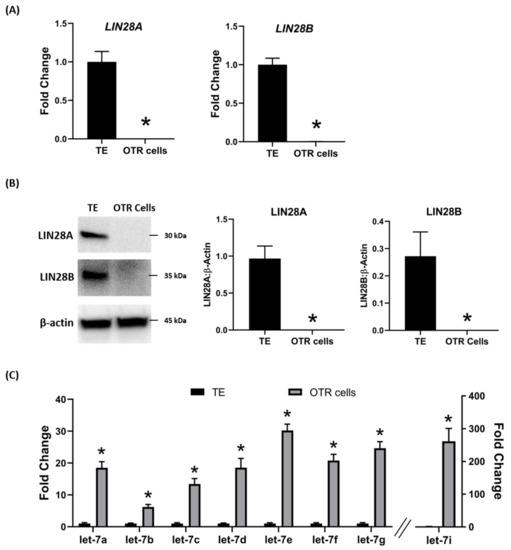
Figure 5.
let-7
A
LIN28A
LIN28B
B
C
Let-7
n
p < 0.05 vs. TE.
The effect of low LIN28 and high
let-7
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC
let-7 miRNAs led to reduced expression of proliferation-associated genes in OTR cells.
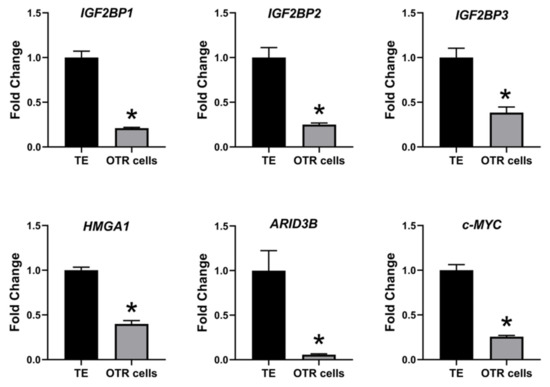
Figure 6.
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC
n
p < 0.05 vs. TE.
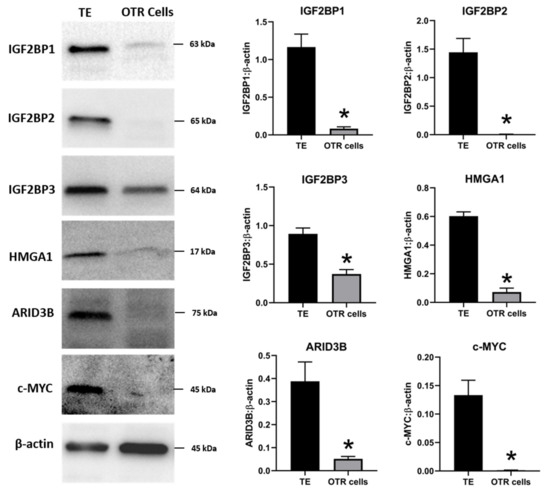
Figure 7.
n
p < 0.05 vs. TE.
The OTR cells originated from day 16 TE undergo senescence after only a few passages. Therefore, the OTR cells were immortalized by overexpressing human telomerase reverse transcriptase (hTERT) to keep them growing for further in vitro experiments. The newly generated immortalized cells were referred to as immortalized ovine trophoblast (iOTR) cells.
3. Overexpression of LIN28 in iOTR Cells Resulted in Increased Expression of Proliferation-Associated Genes
To determine if LIN28 overexpression will rescue the expression of proliferation-associated genes, the iOTR cells were infected with lentiviral particles to generate LIN28A knockin (AKI) or LIN28B knockin (BKI), or lentiviral particles with empty expression vector as expression vector control (EVC). Real-time PCR data showed that AKI iOTR cells had a significant increase in
LIN28A
LIN28B
let-7
let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i
let-7 miRNAs.
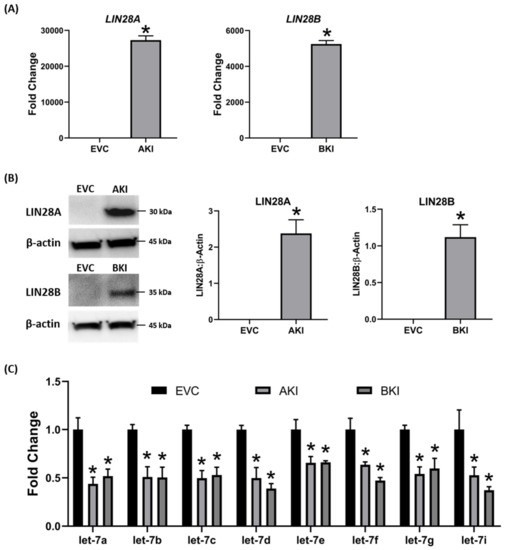
Figure 8.
let-7
A
LIN28A
LIN28B
B
C
Let-7
n
p < 0.05 vs. EVC.
To determine the effect of LIN28 overexpression and reduction in
let-7
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC
let-7 axis.
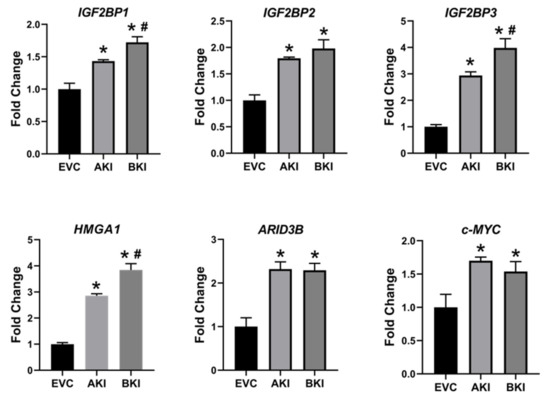
Figure 9.
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC
n
p
p < 0.05 vs AKI.
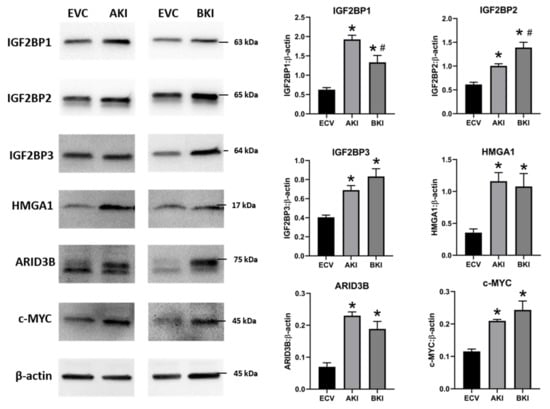
Figure 10.
n
p
p < 0.05 vs AKI.
4. Overexpression of LIN28 Led to Significant Increase in Trophoblast Cell Proliferation
The role of LIN28-
let-7 miRNA axis on the functionality of iOTR cells was determined by measuring proliferation of AKI and BKI iOTR cells compared to EVC after 4 h, 24 h, 48 h, and 72 h. The results showed that proliferation of both AKI and BKI iOTR cells was significantly increased at 24 h, 48 h, and 72 h compared to EVC (Figure 11A). Furthermore, proliferation of BKI iOTR cells was not different at 24 h but was significantly higher at 48 h and 72 h compared to AKI iOTR cells (Figure 11A). The matrigel invasion assay showed that there was no significant change in the invasion index of AKI and BKI iOTR cells compared to EVC (Figure 11B). These results suggest that increased proliferation of AKI and BKI iOTR cells is due to increased expression of proliferation-associated genes in these cells compared to EVC.
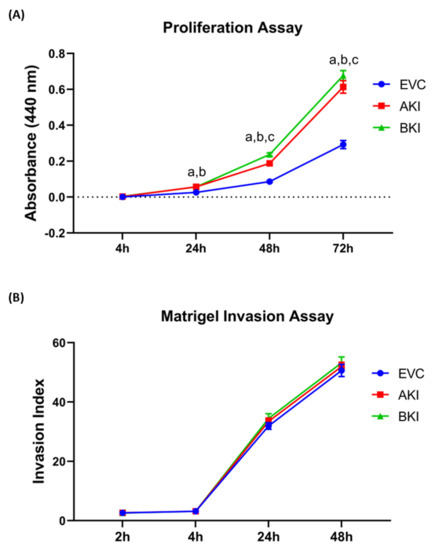
Figure 11.
A
n
B
n
p
p
p < 0.05 for BKI vs. AKI.
5. Discussion
The pluripotency factors LIN28A and LIN28B inhibit the maturation of
let-7 miRNAs [1][2]. Recently, we showed that both Lin28A and LIN28B were significantly decreased and levels of
let-7
let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i) were significantly increased in term human placentas from IUGR pregnancies compared to control pregnancies [3]. We further demonstrated that double knockout of LIN28A and LIN28B in immortalized first trimester human trophoblast (ACH-3P) cells resulted in more robust increase in
let-7
let-7 miRNAs compared to knockin of either LIN28A or LIN28B [3]. In this study, we show that RNA interference (RNAi) of LIN28A or LIN28B in sheep TE in vivo resulted in significant increase in
let-7
let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i). Moreover, the conceptus elongation was significantly reduced after RNAi of LIN28A or LIN28B in TE.
Although animal models with global gene knockout or knockdown have been extensively and successfully used in many studies, it is difficult to exclude the effect of global gene manipulation while focusing on mechanisms involved in one tissue type or organ. Incubating the day 9-hatched sheep blastocyst with shRNA expressing lentiviral particles caused viral infection of only trophoblast cells while all other cells including the inner cell mass were spared of lentiviral infection [4]. Hence, significant reduction in LIN28A or LIN28B in day 16 TE conceptus was restricted to the trophoblast cells only. Conceptus elongation in sheep was due to rapid proliferation of trophoblast cells [5][6][7]; therefore, a significant reduction in conceptus length at day 16 after trophectoderm-specific LIN28A or LIN28B knockdown indicates reduced proliferation of trophoblast cells.
LIN28 is a part of a complex genetic pathway known to regulate multiple downstream targets [8]. Inhibition of biogenesis of mature
let-7 miRNAs is one of the main pathways through which LIN28 regulates the expression of its downstream targets [9][10].
Let-7 miRNAs reduce expression of many genes by degrading their mRNAs or inhibiting translation [1]. Our results show that RNAi of LIN28A or LIN28B and resultant increase in
let-7 miRNAs in day 16 TE significantly reduced IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B, and c-MYC. Previous studies have shown the role of IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B, and c-MYC in cell proliferation [1][11][12][13][14][15][16][17][18], suggesting that reduced proliferation of trophoblast cells after LIN28A or LIN28B knockdown in TE is due to reduced expression of proliferation-associated genes (Figure 12).
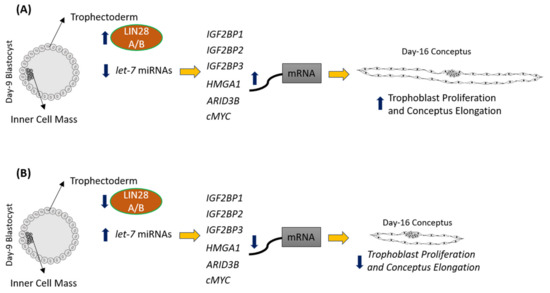
Figure 12.
A
let-7
B
let-7
IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B,
c-MYC, leading to reduced expression of these genes, leading to reduced proliferation of trophoblast cells and, hence, reduced conceptus elongation.
To further investigate the role of the LIN28-
let-7
let-7
let-7 miRNAs suggests that these cells are a differentiated phenotype of trophoblast cells compared to day 16 TE. Furthermore, the expression of proliferation-associated genes including IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B, and c-MYC was also significantly reduced in OTR cells compared to day 16 TE. To overcome the OTR cell senescence, we generated immortalized ovine trophoblast (iOTR) cells by expressing hTERT in these cells. The OTR cells were differentiated and depleted LIN28 and immortalization with hTERT expression did not change LIN28 expression in iOTR cells. Immortalized human first trimester trophoblast cells (Sw.71 cells) were also generated by expressing hTERT in first trimester human trophoblast cells at passage 3 [19]. Sw.71 cells also had depleted LIN28A and LIN28B and high
let-7 miRNAs compared to other trophoblast-derived cell lines such as ACH-3P cells [3]. The iOTR cells generated in this study were similar to Sw.71 cells in terms of LIN28 and
let-7
let-7
let-7 miRNA target genes between day 16 TE and iOTR cells should be taken in consideration if using these cells for further studies.
To generate iOTR cells that were more similar to day 16 TE, we overexpressed LIN28A or LIN28B in iOTR cells. Overexpression of LIN28A or LIN28B in iOTR cells led to a significant decrease in
let-7 miRNAs and significant increase in expression of IGF2BP1, IGF2BP2, IGF2BP3, HMGA1, ARID3B, and c-MYC compared to control. The proliferation of both AKI and BKI iOTR cells was significantly increased compared to control; however, there was no change in cell invasion. We recently showed that knockout of LIN28A and LIN28B in ACH-3P cells reduced cell proliferation but did not affect cell invasion [20]. Both ACH-3P and iOTR cells have low invasion; therefore, it would be hard to see a further reduction in invasion. Moreover, cell proliferation and invasion are two distinct and critical processes during placental development. Reduced trophoblast cell proliferation without reduced invasion can be sufficient to cause impaired conceptus elongation and attachment. These results suggest that the LIN28-
let-7 miRNA axis plays a role in proliferation of immortalized trophoblast cells by regulating the expression of genes associated with cell proliferation. We suggest that the iOTR cells overexpressing LIN28A and LIN28B would be a better choice to study molecular mechanisms in ovine trophoblast cells compared to iOTR cells with depleted LIN28A and LIN28B.
To our knowledge, this is the first in vivo study defining the role of LIN28-
let
let-7
let-7
let-7 miRNAs [3]. MicroRNAs can be easily measured in tissue biopsies, blood, and other biological samples. Based on our studies, we suggest that low LIN28 or high
let-7 miRNAs in placenta could be detected in blood and be used as potential biomarkers for intrauterine growth restriction.
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of Let-7–Regulated Oncofetal Genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef]
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013, 70, 2657–2675. [Google Scholar] [CrossRef]
- JnBaptiste, C.K.; Gurtan, A.M.; Thai, K.K.; Lu, V.; Bhutkar, A.; Su, M.-J.; Rotem, A.; Jacks, T.; Sharp, P.A. Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1–3 family. Genes Dev. 2017, 31, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Degrauwe, N.; Suvà, M.-L.; Janiszewska, M.; Riggi, N.; Stamenkovic, I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Lu, D.; Liu, L.; Cai, J.; Zhou, Y.; Yang, Y.; Zhang, Y.; Zhang, J. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget 2017, 8, 93672–93687. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Francia, G.; Zhang, J.-Y. p62/IMP2 stimulates cell migration and reduces cell adhesion in breast cancer. Oncotarget 2015, 6, 32656–32668. [Google Scholar] [CrossRef]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef]
- Zhou, Y.; Meng, X.; Chen, S.; Li, W.; Li, D.; Singer, R.; Gu, W. IMP1 regulates UCA1-mediated cell invasion through facilitating UCA1 decay and decreasing the sponge effect of UCA1 for miR-122-5p. Breast Cancer Res. BCR 2018, 20, 32. [Google Scholar] [CrossRef]
- Schmiedel, D.; Tai, J.; Yamin, R.; Berhani, O.; Bauman, Y.; Mandelboim, O. The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. eLife 2016, 5, e13426. [Google Scholar] [CrossRef]
- Lederer, M.; Bley, N.; Schleifer, C.; Hüttelmaier, S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin. Cancer Biol. 2014, 29, 3–12. [Google Scholar] [CrossRef]
- Wu, C.; Ma, H.; Qi, G.; Chen, F.; Chu, J. Insulin-like growth factor II mRNA-binding protein 3 promotes cell proliferation, migration and invasion in human glioblastoma. Onco. Targets Ther. 2019, 12, 3661–3670. [Google Scholar] [CrossRef]
- Wan, B.-S.; Cheng, M.; Zhang, L. Insulin-like growth factor 2 mRNA-binding protein 1 promotes cell proliferation via activation of AKT and is directly targeted by microRNA-494 in pancreatic cancer. World J. Gastroenterol. 2019, 25, 6063–6076. [Google Scholar] [CrossRef]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Available online: https://www.hindawi.com/journals/sci/2018/4217259/ (accessed on 18 November 2019).
- Brooks, K.; Burns, G.W.; Moraes, J.G.N.; Spencer, T.E. Analysis of the Uterine Epithelial and Conceptus Transcriptome and Luminal Fluid Proteome During the Peri-Implantation Period of Pregnancy in Sheep. Biol. Reprod. 2016, 95, 88. [Google Scholar] [CrossRef] [PubMed]
- Uchikura, Y.; Matsubara, K.; Muto, Y.; Matsubara, Y.; Fujioka, T.; Matsumoto, T.; Sugiyama, T. Extranuclear Translocation of High-Mobility Group A1 Reduces the Invasion of Extravillous Trophoblasts Involved in the Pathogenesis of Preeclampsia: New Aspect of High-Mobility Group A1. Reprod. Sci. Thousand Oaks Calif. 2017, 24, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol. Cell. Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef] [PubMed]
- Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Cowden Dahl, K.D. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PlLoS ONE 2015, 10, e0131961. [Google Scholar] [CrossRef]
- Roy, L.; Samyesudhas, S.J.; Carrasco, M.; Li, J.; Joseph, S.; Dahl, R.; Cowden Dahl, K.D. ARID3B increases ovarian tumor burden and is associated with a cancer stem cell gene signature. Oncotarget 2014, 5, 8355–8366. [Google Scholar] [CrossRef]
- Ratliff, M.L.; Mishra, M.; Frank, M.B.; Guthridge, J.M.; Webb, C.F. The Transcription Factor ARID3a Is Important for In Vitro Differentiation of Human Hematopoietic Progenitors. J. Immunol. Baltim. Md 1950 2016, 196, 614–623. [Google Scholar] [CrossRef]
- Habir, K.; Aeinehband, S.; Wermeling, F.; Malin, S. A Role for the Transcription Factor Arid3a in Mouse B2 Lymphocyte Expansion and Peritoneal B1a Generation. Front. Immunol. 2017, 8, 1387. [Google Scholar] [CrossRef]
- Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2017, 422, 83–91. [Google Scholar] [CrossRef]
- Lala, N.; Girish, G.V.; Cloutier-Bosworth, A.; Lala, P.K. Mechanisms in decorin regulation of vascular endothelial growth factor-induced human trophoblast migration and acquisition of endothelial phenotype. Biol. Reprod. 2012, 87, 59. [Google Scholar] [CrossRef]
- Baker, C.M.; Goetzmann, L.N.; Cantlon, J.D.; Jeckel, K.M.; Winger, Q.A.; Anthony, R.V. Development of ovine chorionic somatomammotropin hormone-deficient pregnancies. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R837–R846. [Google Scholar] [CrossRef]
- Jeckel, K.J.; Boyarko, A.C.; Bouma, G.J.; Winger, Q.A.; Anthony, R.V. Chorionic Somatomammotropin Impacts Early Fetal Growth and Placental Gene Expression. J. Endocrinol. 2018, 237, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.H.; Cantlon, J.D.; Wright, C.D.; Henkes, L.E.; Seidel, G.E.; Anthony, R.V. The Involvement of Proline-Rich 15 in Early Conceptus Development in Sheep. Biol. Reprod. 2009, 81, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Tsialikas, J.; Romer-Seibert, J. LIN28: roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-L.; Chen, P.-S.; Johansson, G.; Kuo, M.-L. Function and regulation of let-7 family microRNAs. MicroRNA Shariqah United Arab Emir. 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Straszewski-Chavez, S.L.; Abrahams, V.M.; Alvero, A.B.; Aldo, P.B.; Ma, Y.; Guller, S.; Romero, R.; Mor, G. The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 2009, 30, 939–948. [Google Scholar] [CrossRef]
- Hayer, A.; Shao, L.; Chung, M.; Joubert, L.-M.; Yang, H.W.; Tsai, F.-C.; Bisaria, A.; Betzig, E.; Meyer, T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 2016, 18, 1311–1323. [Google Scholar] [CrossRef]
