You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Alessandra Baracca and Version 2 by Jason Zhu.
Mitochondrial ATP synthase (Complex V) catalyzes the last step of oxidative phosphorylation and provides most of the energy (ATP) required by human cells. The mitochondrial genes MT-ATP6 and MT-ATP8 encode two subunits of the multi-subunit Complex V. Since the discovery of the first MT-ATP6 variant in the year 1990 as the cause of Neuropathy, Ataxia, and Retinitis Pigmentosa (NARP) syndrome, a large and continuously increasing number of inborn variants in the MT-ATP6 and MT-ATP8 genes have been identified as pathogenic. Variants in these genes correlate with various clinical phenotypes, which include several neurodegenerative and multisystemic disorders.
- mitochondria
- ATP synthase
- ATP6
- ATP8
- mt-DNA
1. Introduction
The mitochondrial genome presents some specific features, including maternal inheritance and heteroplasmy. Heteroplasmy is a condition in which at least two different mitochondrial genomes are present within the same cell. Pathogenic variants in the mt-DNA are highly recessive and usually coexist with the wild-type mt-DNA molecules. Therefore, the clinical manifestation of mt-DNA variants mainly depends on both their severity and the mutational load (heteroplasmy) of the tissues [1][2][2,8].
Over the last two decades, a large number of studies using different patient’s specimens and other cellular paradigms have led to in-depth investigations of the biochemical and cellular alterations caused by MT-ATP6 and MT-ATP8 variants, which are summarized in Table 1.
As shown in Table 1, the cellular dysfunctions observed for a given variant can be highly variable, and one element that contributes to this characteristic is heteroplasmy.
Consequently, as with other mt-DNA-associated diseases, a specific feature is the threshold of the percentage of mutant genome (or percentage of heteroplasmy) that must be exceeded to detect a biochemical alteration. As reported below, this defect may also depend on the variant, haplogroup, cell type, and tissue type.
Table 1. Biochemical and cellular parameters in patient tissues and cell models carrying MT-ATP6 and MT-ATP8 pathogenic variants. Abbreviation: heteroplasmy (H), oxygen consumption rate (OCR), mitochondrial membrane potential (MMP), reactive oxygen species (ROS), induced pluripotent stem cell (iPSC) and neural progenitor cells (NPCs), Normal (N), Decreased (D), Increased (I), Affected (A).
| Genetic Variant/Subunit AA Change | Tissue or Cell Models H (%) |
Biochemical and Cellular Parameters | ||||
|---|---|---|---|---|---|---|
| CV ATP Synthesis |
CV ATP Hydrolysis |
OCR | CV Assembly/Stability | Other Mitochondrial and Cellular Readouts |
||
| m.8382C>T ATP8: p.T6I |
Muscle (100%) [3][34] |
(D) | CI activity (D) | |||
| Fibroblasts (100%) [3][34] |
(N) | (N) | CIV activity (D) | |||
| m.8403T>C ATP8: p.I13T |
Fibroblasts (100%) |
(N) [3][34] | (N) [3][34] | Depolarized plasma membrane and ROS (I) [4][35]; CIV activity (D) [3][34] |
||
| Yeast (100%) [5][36] |
(D) | (N) | Growth in stress conditions (D); Mitochondrial membrane potential (N) |
|||
| m.8424T>C ATP8: p.I20P |
Muscle (100%) [3][34] |
(D) | CI, CII, CIII, and CIV activities (D) | |||
| Fibroblasts (100%) [3][34] |
(D) | (N) | CI activity and growth in galactose media (D) | |||
| Cybrids (100%) [3][34] |
(D) | (D) | CI and CIV activities (D); Lactate production (I) |
|||
| m.8528T>C ATP8: p.W55R ATP6: p.M1T |
Fibroblast (93%) [6][37] |
(D) | ||||
| Heart muscle (90%) [7][38] | (A) | CV subunit levels and CI activity (D); ATP6 and ATP8 protein levels (D) |
||||
| m.8529G>A ATP8: p.W55X ATP6: p.M1M |
Muscle (>90%) [8][39] |
(D) | (D) | (A) | CI-CIV activities (N) | |
| Fibroblast (>90%) [8][39] |
(D) | CI-CIV activities (N) | ||||
| Cybrids (100%) |
(D) [8][9][39,40] | (D) [9][40] | (A) [8][9][39,40] | Growth in galactose media (D); ATP6 and ATP8 protein levels (D); Complexes II, III, and IV levels (D) [9][40] |
||
| m.8561C>G ATP8: p.P66A ATP6: p.P12R |
Myoblasts (99%) [10][41] |
(A) | Total ATP level (D); ROS (N); ATP6 and ATP8 protein levels (N) |
|||
| m.8561C>T ATP8: p.P66L ATP6: p.P12S |
Muscle (99%) [11][42] | (D) | (A) | |||
| m.8611insC ATP6: p.L29PfsX36 |
Muscle (60%) [12][43] |
(D) | (A) | |||
| Fibroblasts (80%) [12][43] |
(D) | (A) | ATP6 protein level (D); Mitochondrial cristae structure and dynamics (A) |
|||
| m.8618insT ATP6: p.T33HfsX32 |
Muscle (65–85%) |
(A) [13][14][44,45] | ATP6 protein level (D) [13][44] | |||
| Fibroblasts (45%) [14][45] |
(D) | (A) | ROS (I); Mitochondrial network morphology (N) |
|||
| m.8648G>A ATP6: p.R41Q |
Fibroblast (100%) [3][34] |
(N) | (N) | |||
| Cybrids (100%) [3][34] |
(N) | (N) | ||||
| m.8782G>A ATP6: p.G86X |
Fibroblasts (12–27%) [14][45] |
(D) | (A) | ROS (I); Mitochondrial morphology (N) |
||
| m.8806C>G ATP6: p.P94A |
Muscle (100%) [3][34] |
(N) | CI-CIV activities (D) | |||
| m.8839G>C ATP6: p.A105P |
Cybrids (100%) [15][46] |
(N) | Growth in galactose media (D); Mt-DNA copy number (I); OXPHOS protein levels (I); Mitochondrial membrane potential (D); CI-CIV activities (N) |
|||
| m.8843T>C ATP6: p.I106T |
Yeast (100%) [16][47] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.8851T>C ATP6: p.W109R |
Yeast (100%) [17][48] |
(D) | (D) | (D) | (N) | Growth in stress conditions (D); Mitochondrial cristae structure (A); CIII and CIV super-complexes (D) |
| m.8909T>C ATP6: p.F128S |
Yeast (100%) [18][49] |
(D) | (D) | (A) | ||
| m.8932C>T ATP6: p.P136S |
Yeast (100%) [19][50] |
(D) | (D) | (A) | ATP6 protein level (D) | |
| m.8946A>C ATP6: p.M140I |
Fibroblasts (100%) [3][34] |
(N) | (N) | CI activity (D) | ||
| m.8950G>A ATP6: p.V142I |
Lymphocytes [20][51] | (D) | ||||
| Yeast (100%) [16][47] | (D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
||
| m.8969G>A ATP6: p.S148N |
Muscle (100%) [3][34] | (N) | CI activity (D) | |||
| Yeast (100%) |
(D) [21][22][52,53] | (D) [21][52] | (D) [21][52[22],53] | (A) [21][52] | Growth in stress conditions (D) [21][22][52,53] | |
| Cybrids (19–98%) [21][52] |
(D) | Mitochondrial cristae structure (A); ROS (I) |
||||
| Fibroblasts (100%) [23][54] |
(D) | |||||
| m.8975T>C ATP6: p.L150P |
Muscle [3][34] | (D) | CI activity (D) | |||
| Fibroblasts (100%) [3][34] |
(D) | (N) | CI activity (D); Growth in galactose media (D) |
|||
| Cybrids (100%) [3][34] |
(N) | (N) | CI and CIV activities (D); Growth in galactose media (D); Lactate production (I) |
|||
| m.8989G>C ATP6: p.A155P |
Muscle (92%) [24][55] |
(D) | Mitochondrial ultrastructure (N) | |||
| m.8993T>G ATP6: p.L156R |
Yeast (100%) [25][56] |
(D) | (D) | (D) | (A) | Growth in stress conditions (D); CIV level (D) |
| Platelets (80–93%) |
(D) [26][27][57,58] | (N) [26][27][57,58] | CV ATP-driven proton flow (N) [26][57] | |||
| Lymphocytes (80–100%) |
(D) [28][29][30][31][59,60,61,62] | (D) [28][59] | (D) [29][60] | ROS and mitochondrial membrane potential (I) [31][62]; CV proton flow (D) [30][32][61,63]; Oligomycin sensitivity of CV proton flow (I) [32][63] |
||
| Muscle (76%) [33][64] |
(A) | |||||
| Fibroblasts (70–100%) |
(D) [3][34][35][36][37][38][39][34,65,66,67,68,69,70] | (D) [35][38][40][66,69,71]; (N) [3][34][36][34,65,67] | (D) [41][42][72,73]; (N) [34][65] |
(N) [36][37][67,68] | Mitochondrial membrane potential (I) [36][39][67,70]; Mitochondrial morphology (A) [39][42][70,73]; ROS (I) [39][43][70,74]; Antioxidant enzymes (A) [39][70]; Oligomycin sensitivity of CV (I) [34][65]; Growth in galactose media (D) [3][35][38][40][34,66,69,71]; Mitochondrial calcium uptake (D) [39][70]; Glycolytic capacity (D) [42][73]; CI and CIV activities (D) [3][34] |
|
| Cybrids (45–100%) |
(D) [3][35][37][40][44][45][,6646,68],71,75[,7647][48][49][34,77,78,79,80] | (D) [3][34]; (N) [40][71] |
(D) [29]60[40][46][47][,7148,77],78[50,79][51][,81,82] | (A) [37][44][48][68,75,79] | Mitochondrial membrane potential (D) [50][81] or (I) [47][49][78,80]; Mitochondrial morphology (A) [52][53][83,84]; Mitochondrial ultrastructure (A) [49][80]; ROS (I) [47][50][54][78,81,85]; Antioxidant enzymes (A) [47][54][78,85]; Growth in galactose media (D) [35][45][66[47],76,78]; ATP level (D) [50][81]; Extracellular lactate (I) [3][46][34,77]; Autophagy (I) [53][84]; CI, CII, or CIV activities (D) [3][47][48][34,78,79]; Oligomycin [37][68] and apoptosis [49][80] sensitivity (I); Actin cytoskeleton and Ca2+ in-flux rates (A) [52][83]; Reductive carboxylation of glutamine and NADH/NAD ratio (I) [51][55][82,86] |
|
| IPSCs (90–100%) |
(D) [56][87]; (N) [57][88] | Mitochondrial membrane potential, ROS, and lactate production (I) [58][89] | ||||
| NPCs, Neurons (90–100%) |
(D) [58][89] | Mitochondrial membrane potential, ROS, and antioxidant enzymes (I) [58][89]; Degenerative defect [58][89]; Metabolic dysregulation; Formation of cerebral organoid (A) [57][88] |
||||
| m.8993T>C ATP6: p.L156P |
Yeast (100%) [59][90] |
(D) | (N) | (D) | (N) | CIV level, COX2, and ATP6 protein levels (D) |
| Lymphocytes (90–95%) |
(D) [31][62] | Mitochondrial membrane potential (N); ROS (I) [31][62]; Proton flux (D) [32][63] |
||||
| Fibroblasts (95–100%) |
(D) [3][34][34,65] | (D) [3][34], (N) [34][65] | (N) [34][65] | (N) [37][68] | Depolarized plasma membrane and ROS (I) [4][35]; Growth in galactose media (D) [3][34] |
|
| Cybrids (100%) |
(D) [40][46][71,77]; (N) [3][34[48],79] |
(N) [3][34] | (D) [46][77]; (N) [48][79] |
(N) [37][48][68,79] | Lactate production (I) [3][46][34,77] | |
| m.9008C>G ATP6: p.T161S |
Muscle (100%) [3][34] |
(N) | ||||
| Fibroblasts (100%) [3][34] |
(N) | (N) | CI activity (D) | |||
| Cybrids (100%) [3][34] |
(D) | (N) | Growth in galactose media (D); Lactate production (I) |
|||
| m.9016A>G ATP6: p.I164V |
Yeast (100%) [16][47] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9019A>G ATP6: p.T165A |
Muscle (100%) [3][34] |
(D) | CI activity (D) | |||
| m.9025G>A ATP6: p.G167S |
Yeast (100%) [16][47] |
(D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
|
| m.9029A>G ATP6: p.H168R |
Yeast (100%) [16][47] |
(D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
|
| Cybrids (100%) [50][81] |
(D) | ATP level (D); ROS and mitochondrial membrane potential (I) |
||||
| m.9032T>C ATP6: p.L169P |
Cybrids (25–80%) [50][81] |
(D) | ATP level (D); ROS and mitochondrial membrane potential (I) |
|||
| m.9035T>C ATP6: p.L170P |
Cybrids (100%) [60][91] |
(D) | ROS and antioxidant enzymes (I); Mitochondrial membrane potential (N); Sensitivity to glucose deprivation (I); Oxidative stress (I) |
|||
| Muscle (100%) [3][34] |
(D) | CI activity (D) | ||||
| Fibroblasts (100%) |
(D) [3][34] | (N) [3][34] | (D) [61][92] | (A) [61][92] | Growth in galactose media (D) [3][34] | |
| m.9058A>G ATP6: p.T178A |
Yeast (100%) [16][47] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9101T>C ATP6: p.I192T |
Lymphocytes (100%) [62][63][93,94] |
(D) | ||||
| Cybrids (100%) [63][94] |
(D) | |||||
| m.9127 delAT ATP6: p.I201PfsX2 |
Fibroblasts (50%) [64][95] |
(D) | (D) | (N) | Oligomycin-induced increase in mitochondrial membrane potential (D) | |
| m.9134A>G ATP6: p.E203G |
Muscle [65][96] | (D) | (D) | |||
| m.9139G>A ATP6: p.A205T |
Yeast (100%) [16][47] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9154C>T ATP6: p.Q210X |
Fibroblasts [66][97] | (N) | (A) | Mitochondrial morphology (A) | ||
| IPSC and Neurons [66][97] | (A) | Motor neuron differentiation (A); Mitochondrial morphology (A); Hyperactivation of the Notch pathway |
||||
| m.9160T>C ATP6: p.Y212H |
Yeast (100%) [16][47] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9176T>G ATP6: p.L217R |
Yeast (100%) [67][98] |
(D) | (D) | (A) | Growth in stress conditions (D); CIV super-complexes (D); ATP6, COX2, and CYTB protein levels (D); Mitochondrial ultrastrucure (A) |
|
| Muscle (>95%) [33][64] |
(A) | |||||
| Fibroblasts (95–100%) |
(D) [68][99] | (N) [40][71] | (N) [68][99] | Mitochondrial membrane potential (I) [68][99]; Growth in galactose media (D) [40][71] |
||
| Cybrids (30–100%) |
(D) [40][48][49][71,79,80] | (N) [40][71] | (D) [40][48][71,79] | (A) [48][79] | CI and CIV activities (D) [48][79]; Mitochondrial ultrastructure (A) [49][80]; Mitochondrial membrane potential and apoptosis sensitivity (I) [49][80] |
|
| m.9176T>C ATP6: p.L217P |
Yeast [69][100] | (D) | (N) | (D) | (A) | |
| Muscle (100%) [3][34] |
(D) | |||||
| Fibroblasts (100%) |
(N) [70][101]; (D) [71][102] | (N) [40][71] | (A) [71][102] | Mitochondrial network morphology (N) [71][102]; Depolarized plasma membrane and ROS (I) [4][35] |
||
| Cybrids (100%) [40][71] |
(D) | (N) | ||||
| m.9185T>C ATP6: p.L220P |
Yeast [72][103] | (D) | (N) | (N) | (N) | Sensitivity of growth to oligomycin (I) |
| Muscle (>97%) |
(D) [73][74][104,105] | (A) [74][105] | CI, CII, and CIV activities (N) [75][76][77][106,107,108] | |||
| Lymphocytes [75][106] |
(N) | (D) | ||||
| Fibroblasts (90–100%) |
(D) [3][4][34,35] | (D) [4][35]; (N) [3][34] | (D) [4][35] | (N) [4][35] | CI activity (D) and depolarized plasma membrane [4][35]; ROS or antioxidant enzymes (I) [4][78][35,109]; CI, CII, and CIV activities (N) [75][106]; Mitochondrial membrane potential (N) [78][109]; Lactate production (I) [3][34] |
|
| Cybrids (100%) | (D) [78][109]; (N) [3][34] |
(D) [4][35]; (N) [3][34] | (D) [4][35] | CI activity (D) [4][35]; Lactate production (I) [3][34]; Mitochondrial membrane potential (N) [78][109] |
||
| NPC and neuron (100%) [78][109] | (D) | (N) | Mitochondrial membrane potential (I); Mitochondrial calcium homeostasis (A); Depolarized plasma membrane; Mitochondrial cristae structure and ROS (N) |
|||
| m.9191T>C ATP6: p.L222P |
Muscle (94%) [73][104] |
(D) | (D) | |||
| Yeast [72][79][103,110] |
(D) | (N) | (D) | (A) | Growth in stress conditions (D); CIV level (D); ATP6 protein level (D) |
|
| m.9205delTA ATP6: p.X227NA |
Muscle (>98%) [80][111] |
CIV activity (D) | ||||
| Fibroblasts (>98%) |
(D) [80][111] | (N) [80][111] | (D) [80][111] | (A) [80][111] | CIV activity (D) [80][81][111,112]; ATP6 protein and CIV subunit levels (D) [80][111]; Morphological abnormalities of mitochondria [81][112] |
|
2. The mt-DNA Pathogenic Variants at Position m.8993
The two most common variants in MT-ATP6 are m.8993T>G (p.Leu156Arg) and m.8993T>C (p.Leu156Pro), which cause a change in a highly conserved leucine residue on ATP6 [26][82][83][13,57,113]. These variants are the most common and are responsible for approximately 50% of reported MT-ATP6 disease cases [84][33]. These variants are associated with Neuropathy, Ataxia, and Retinitis Pigmentosa (NARP) or Maternal Inherited Leigh syndrome (MILS) when heteroplasmy is between 70 and 90% or greater than 90%, respectively. Furthermore, the T>G transversion usually results in a more severe clinical phenotype than the T>C transition [82][83][85][86][13,113,114,115].
2.1. Biochemical and Cellular Dysfunctions in Mutated Cell Models
Analyses of patient specimens carrying the m.8993T>G or the m.8993T>C variant have been performed in platelets [26][27][57,58], lymphocytes [28][29][30][31][32][59,60,61,62,63], muscle tissue [33][64], and skin fibroblasts [3][4][34][35][36][37][38][39][40][41][42][43][87][34,35,65,66,67,68,69,70,71,72,73,74,116] (see Table 1). Several biochemical abnormalities have been identified, including a decreased ATP synthesis and oxygen consumption rate (OCR) [3][26][27][28][29][30][31][34][35][36][37][38][39][40][41][42][34,57,58,59,60,61,62,65,66,67,68,69,70,71,72,73], often in direct correlation with the mutation load [27][30][40][45][58,61,71,76], an alteration of the proton flux [30][31][32][61,62,63], and a not-fully assembled CV [33][64]. In human cells, these ATP synthase dysfunctions lead, as secondary effects, to a reduction in growth in stress medium [3][35][38][40][34,66,69,71], to an increase in both mitochondrial membrane potential (MMP) [31][36][39][87][62,67,70,116] and ROS [4][31][39][43][35,62,70,74], as well as to an altered mitochondrial network morphology and cristae structure [39][42][70,73].
In cells of NARP patients carrying the m.8993T>G variant, the severe impairment of OXPHOS has been proposed as the primary pathogenic defect; instead, the increase in ROS could be the main contributor to the pathogenesis of the disease associated with the m.8993T>C variant [31][62].
It is worth noting that no significant effects of the m.8993T>G variant have been reported on either ATP hydrolytic activity or ATP-driven proton transport by Complex V in patient cells [26][36][57,67]. However, inhibition of ATP hydrolytic activity could contribute to energy preservation and survival of cells under stress conditions (oxygen shortage), leading to the collapse of the proton motive force and ATP synthase working in reverse. Incidentally, due to the heterogeneity of the membrane potential within the same mitochondrion and the possible coexistence of ATP synthase working physiologically and in reverse [88][89][117,118], patients might benefit from the use of a specific inhibitor of the hydrolytic activity of CV.
The different percentages of heteroplasmy and other factors, including nuclear background and the type of tissue analyzed, may contribute to the phenotypic differences observed in the analysis of patients’ specimens, as well as those observed in clinical outcomes [61][84][90][33,92,119]. For these reasons, transmitochondrial cybrids are widely used to validate the possible pathogenicity of a mitochondrial variant, even in homoplasmic populations, with the advantage of using a cell model with the same nuclear background [91][120]. In the case of the two MT-ATP6 variants, this cell model clearly demonstrated the impairment of respiration [29][40][46][47][48][50][51][60,71,77,78,79,81,82], ATP synthesis [3][35][37][40][44][45][46][47][48][49][34,66,68,71,75,76,77,78,79,80], mitochondrial morphology [52][53][83,84], and enhanced ROS production [47][50][54][78,81,85], confirming the milder effect of m.8993T>C compared to the T>G variant [31][37][40][46][48][62,68,71,77,79].
Interestingly, analysis of different cybrid lines carrying the same m.8993T>G variant highlighted that the mitochondrial genome sequence, and thus the haplogroup, is a factor contributing to the variations in the observed biochemical phenotypes, ranging from normal to severe defects [48][79]. Moreover, clear evidence of the role of mutation load on deleterious biochemical abnormalities has been recently reported in isogenic cybrids, where the OCR reduction and the extracellular acidification rate (ECAR) increase were proportionally linked to the levels of heteroplasmy, indicating a switch toward glycolysis [51][82]. Metabolic remodeling induced by the m.8993T>G variant was also investigated, and both proteomics and metabolomics analysis were consistent with increased glycolysis and reductive carboxylation of glutamine to support cell survival and to maintain redox balance [51][82]. Accordingly, a second report showed that, in cybrids, the impaired OXPHOS activity induces compensatory energy-generating anaplerotic mechanisms where glutamine-glutamate-α-ketoglutarate metabolism sustains cell survival [55][86].
The deleterious mechanism hypothesized based on all these studies, especially for the m.8993T>G variant, includes defective proton transport across Fo, failure of the enzyme to couple phosphorylation of ADP on F1 to proton flow, or alteration of the holoenzyme assembly and stability [30][31][32][84][33,61,62,63]. Considering that these alterations have been observed in different cellular models despite not always being together, it seems reasonable that all three mechanisms contribute to the pathogenicity of the variants.
In recent years, the introduction of induced pluripotent stem cell (iPSC) technology allowed disease modeling by overcoming the difficulty of accessing clinically relevant patients’ cells or tissues, such as neurons. The generation of IPSCs requires multiple quality checks and presents the issue of heteroplasmy fluctuations due to the genetic bottleneck occurring in the reprogramming process. In addition, the mutant load can also change during differentiation or cell culture and therefore must be constantly monitored [92][93][121,122].
A series of patient-derived iPSCs carrying the m.8993T>G or T>C variant has been developed [58][94][95][96][97][89,123,124,125,126], and neural progenitor cells (NPCs) and neurons have been differentiated [58][89]. The mutant iPSCs were able to differentiate into the three embryonic germ layers (endoderm, mesoderm, and ectoderm) [78][96][97][109,125,126]. However, analysis of embryoid bodies showed impaired differentiation potential in cells with a high percentage of the variant [96][125]. Overall, the generated cell types recapitulate the energy defects observed in other cell models and the degenerative phenotypes observed in patients [56][58][87,89]. Neurons, in part because of their predominantly mitochondria-dependent oxidative metabolism [58][78][89,109], have shown degenerative defects not detectable in other, less differentiated cells, notably ATP shortage and AMPK activation, finer neuronal fibers, and increased sensitivity to glutamate toxicity [58][89]. Furthermore, a study of mutant IPSCs revealed abnormalities during the three-dimensional differentiation and a defective formation of cerebral organoids, particularly in the generation of neural epithelial buds, as well as impaired corticogenesis with an altered metabolic profile [57][88].
2.2. Modeling of ATP6 Subunit Carrying Changes in Leu156 in the ATP Synthase Human Structure
In the recently released ATP synthase human structures ([98][16], PDB id 8H9S, 8H9T, 8H9U, and 8H9V for states 1, 2, 3a, and 3b, respectively), the Leu156 residue is located on helix H5 and is buried in the core of ATP6, forming van der Waals contacts with Leu217 and Val218, located on helix H6 (Figure 13). In the four structures, the residues in the vicinity of Leu156 are the same, and there are no conformational transitions involving ATP6 in the different states of the human ATP synthase. The variant of Leu156 in arginine can damage the interaction between subunit helices H5 and H6 because of (i) the larger volume of an arginine residue with respect to a leucine and (ii) the presence of a charged side chain in a region populated only by hydrophobic residues. In other words, the presence of an arginine in position 156 can cause a divarication between helices H5 and H6 that in turn can alter the folding of ATP6. Indeed, the correct positioning of these helices is crucial for proton access to the negatively charged Glu58 in the c subunits (Figure 13).
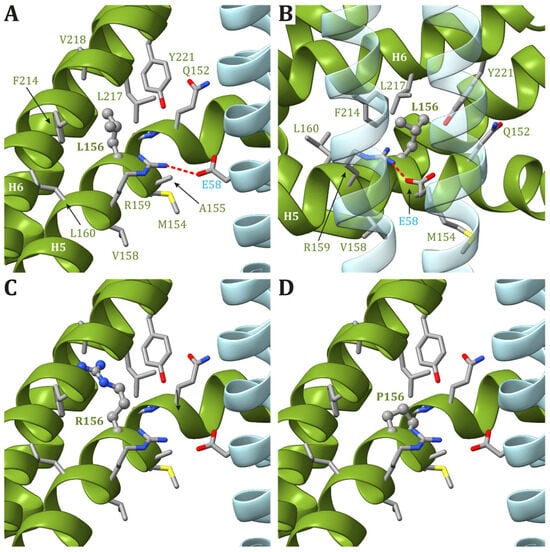
Figure 13. Detail of the region comprising Leu156 of ATP6 in the human structure of ATP synthase (state 1). (A,B) The native ATP6 and c subunits are reported in ribbons colored as in Figure 2. Residues labels are colored as the corresponding subunits. Leu156 is in ball-and-stick, while other residues in the vicinity of Leu156 or proposed to be part of the proton translocation process are in stick. The side chains are colored according to the atom type. The interaction between Arg159 in ATP6 and Glu58 in c8-ring is shown. The orientation of panel (B) is clockwise rotated by 90° around the vertical axis with respect to the orientation in panel (A). Panels (C,D) reports the model structures of the p.Leu156Arg and p.Leu156Pro variants, respectively.
3. The Pathogenic Variants at Nucleotide m.9176
The m.9176T>C and m.9176T>G variants, which cause a Leu217Pro and a Leu217Arg ammino acid change, are frequently found in MILS patients and were first reported in 1995 and 2001, respectively [68][70][99,101].
In the case of the m.9176T>G variant, a partially disassembled CV, decreased ATP synthesis, and mitochondrial respiration due to a defective OXPHOS pathway led to an increase in MMP in MILS patient-derived fibroblasts [40][68][71,99]. Similar observations have been described in cybrids [40][48][49][71,79,80] and in patient-derived muscle tissue [33][64]. Furthermore, analysis of this variant in yeast highlighted a severe reduction in the ATP6 protein level, suggesting that it may affect the assembly of the ATP synthase complex and cause mitochondrial and bioenergetic dysfunctions [67][98]. Human IPSCs have been recently generated for the m.9176T>G [97][126], and their use to develop neurons will be instrumental in deeply characterizing the pathogenic mechanism of this variant in a disease-target tissue.
In the first report in 1995, biochemical analysis of the m.9176T>C variant revealed no defects in ATP synthase function in patient cells with the homoplasmic variant [70][101]. However, further studies reported impaired ATP synthesis [40][69][71][71,100,102] and CV stability [71][102] in both human cells and mutant yeast. As for the variants at the nucleotide m.8993, the alteration caused by the T>C variant was less severe than that caused by the T>G variant [40][69][71,100].
In human structures, Leu217 is positioned in helix H6 of ATP6 and is part of both the interface between helices H5 and H6 and between ATP6 and the c8-ring. While the residues in ATP6 in the vicinity of Leu217 (Leu156, Arg159, Val213, Leu216, Leu220, and Tyr221) do not change during the catalytic cycle, the residues found close to Leu217 in the c subunit are different depending on the ATP synthase state. Indeed, in state 1, Leu52 and Leu56 in the c subunit are close to Leu217 in ATP6 (Figure 24A), while in states 2 and 3a, Leu217 is in the vicinity of Leu52 and Ala55 in the c subunit (Figure 24B). Finally, in state 3b, no residue from any c subunit is in the closeness of Leu217 (Figure 24C). The variant of Leu217 in an arginine residue appears to cause two effects: (i) arginine is a larger residue with respect to leucine, causing some sort of friction between the ATP6 and the c8-ring that is rotating during the catalytic cycle, and (ii) the arginine has a charged side chain that can form H-bonds with other residues in the vicinity, such as Tyr221 from ATP6. This newly formed H-bond can interfere with the formation of another H-bond between Tyr221 and a water molecule in the outlet proton translocation half-channel. The latter water molecule is held in the correct position by two H-bonds, the already cited one with Tyr221 and a second with Glu58 from the c subunit. On the other hand, the variant of Leu217 in a proline residue can cause some sort of effects on the folding of helix H6 downstream of the mutated residue, but—as in the case of p.Leu156Pro—the damage caused by the presence of a small hydrophobic residue in position 156 should be moderate.
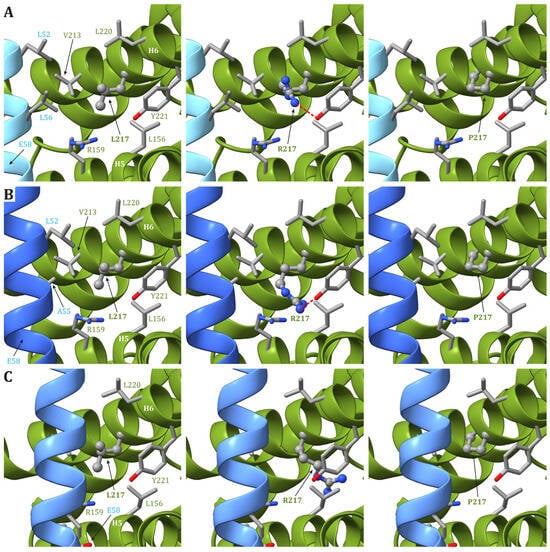
Figure 24. Detail of the region comprising Leu217 from ATP6 in the human structure of ATP synthase in state 1 (A), state 2 and 3a (B), and 3b (C). The native ATP6 and c subunits are reported in ribbons colored as in Figure 2. Residue labels are colored as the corresponding subunits. Leu217 is shown as ball-and-stick, while other residues in the vicinity of Leu217 or proposed to be part of the proton translocation process are shown as a stick. The side chains are colored according to the atom type. In the left panels, the wild-type protein is reported, while the models of Leu217Arg and Leu217Pro variants are reported in the central and right panels, respectively. H-bonds are indicated using dashed red lines.
4. The mt-DNA Pathogenic Variant at Position m.9185
The variant at nucleotide m.9185T>C (p.Leu220Pro) was first reported in 2005 [73][104], and functional studies revealed a moderate effect of this variant on ATP synthase functioning. Indeed, in patient cells, besides normal Complexes I-IV activity [73][76][77][104,107,108], a slight alteration of CV and a depolarization of the plasma membrane were reported, often only in the case of homoplasmy [3][4][73][74][75][78][34,35,104,105,106,109]. Mild effects on ATPase function have also been observed in mutated cybrids and yeast cells [3][4][72][78][34,35,103,109]. The evaluation of IPSCs and their patient-derived NPCs [78][99][109,127], in addition to the defective ATP production, allowed people to highlight mitochondrial impairment that is hidden in the other cell types. Indeed, neural cells presented a mitochondrial hyperpolarization and an alteration of mitochondrial calcium homeostasis, as evidenced by both transcriptomic and proteomic analysis [78][109]. All these data suggest that the variant may alter the ability of these cells to produce ATP and control MMP, causing neural impairment.
In the human ATP synthase structures, Leu220 is located on helix H6 of ATP6, just one helix turn away from Leu217, and forms van der Waals contacts with Met60, located on helix H3. Except for Glu224, the other residues found in the vicinity of Leu220 in ATP6 are all hydrophobic (Leu216, Leu217, and Tyr221). As for Leu217, Leu220 is at the interface between ATP6 and the c8-ring, and the interacting residues from the latter depend on the ATP synthase state. Leu220 of ATP6 is in the vicinity of some residues of the c subunit: Phe47 and Ile51 in states 1 and 3b (Figure 35A) and Leu52 in states 2 and 3a (Figure 35B). As for the previously discussed cases, a variant of Leu220 in a proline residue can have some effects on the fold of helix H6 downstream of the mutated residue and, in turn, cause some problems to the ATP synthase mechanism. On the other hand, proline is a small hydrophobic residue, and no serious steric or electrostatic effects are expected.
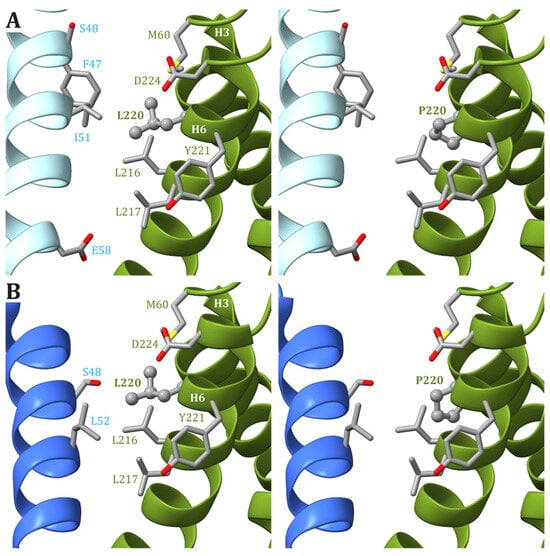
Figure 35. Detail of the region comprising Leu220 from ATP6 in the human structure of ATP synthase in states 1 and 3b (A) and states 2 and 3a (B). The native ATP6 and c subunits are reported in ribbons colored as in Figure 2. Residue labels are colored as the corresponding subunits. Leu220 is shown as ball-and-stick, while other residues in the vicinity of Leu220 or proposed to be part of the proton translocation process are shown as a stick. The side chains are colored according to the atom type. In the left panels, the wild-type protein is reported, while the models of Leu220Pro variants are in the right panels.
5. Other MT-ATP6 and MT-ATP8 Pathogenic Variants
A number of MT-ATP6 and MT-ATP8 variants reported in the literature have been reviewed by two different research groups [82][84][13,33], and the functional studies of cell models are reported in detail in Table 1.
The study of the m.8909T>C variant, found in a patient also carrying the pathogenic m.3243A>G variant in mt-tRNALeu (MT-TL1), reported a defect in Complex V assembly and ATP synthesis [18][49]. A compromised assembly of ATP synthase and a reduced OCR has been observed in fibroblasts of two patients carrying the same m.8782G>A variant, one presenting adult-onset cerebellar ataxia, chronic kidney disease, and diabetes, whereas the other had myoclonic epilepsy and cerebellar ataxia [14][45].
The truncating variant m.9154C>T was found in a patient with adult-onset axonal neuropathy, ataxia, and IgA nephropathy and caused alteration of Complex V assembly, mitochondrial morphology, and ultrastructure in mutated fibroblasts [66][97]. Interestingly, the mutation load resulted to be proportional to Complex V assembly defect in patient-derived iPSCs and responsible for impaired neurogenesis due to Notch hyperactivation and altered metabolism of mature motor neurons [66][97].
Other identified MT-ATP6 variants include the m.8858G>A variant in a sporadic case of NARP-MILS [100][128]; the m.8936T>A in a young boy with atypical mitochondrial Leigh syndrome associated with bilateral basal ganglia calcifications [101][129]; m.9143T>C in a patient with insulin-dependent diabetes mellitus, recurrent lactic acidosis, infections, and immunodeficiency [102][130]; m.9154C>T in a patient with neuropathy, cerebellar ataxia, and IgA nephropathy [103][131]; and m.9171A>G in a patient with mitochondrial retinopathy with atrophy [104][132]. The three variants m.8572G>A, the m.8578C>T and m.8812A>G were found in patients with adult-onset spinocerebellar ataxia (SCA) [105][133].
Regarding new variants affecting MT-ATP6, MT-ATP8, or both, m.8561C>T, which causes a defect in CV assembly, was reported in a child with early onset ataxia, psychomotor delay, and microcephaly [11][42], whereas functional studies have been performed for the m.8382C>T, m.8424T>C, m.8806C >G, m.8975T>C, m.9008C>G, and m.9019A>G variants [3][34].
