Deep learning (DL) has been applied successfully in medical imaging such as reconstruction, classification, segmentation, and detection.
- deep learning
- 3D MRI
- MRI datasets
- DL tools
1. Introduction
2. DL Architectures
Deep neural networks (DNNs) are used for medical-image reconstruction, quality enhancement, feature mapping, contrast transformation, classification of tumors or cancer types, and segmentation for detecting normal and abnormal tissues. Deep architectures can extract features from data in place of conventional hand-crafting feature extraction algorithms. DL can reconstruct high-quality images from undersampled data via discovering complex mappings using undersampled k-space data and fully sampled images. Several DL architectures used for MRI reconstruction are described below. A convolutional neural network (CNN) [23][21] (Figure 1a) is an efficient approach to DNNs that is particularly effective in image processing and computer vision (CV) tasks. It consists of a set of convolutional layers and applies convolution operations to the input data. These operations involve sliding small filters (kernels) over the input image to learn local features. Through these convolution operations, the network captures low-level features (e.g., edges, textures) in the early layers and progressively more abstract and complex features in the deeper layers. The convolutional layers produce feature maps that represent learned patterns and features in the input data. Thus, CNNs automatically learn hierarchical representations of features in images, making them well-suited for tasks related to images and videos. CNNs have been widely successful in tasks such as image reconstruction, classification, object detection, and segmentation. Google, Microsoft, and Facebook have established research groups to examine novel CNN designs [24][22]. A CNN deals with raw images and, in some cases, minimizes the data pre-processing tasks. The AlexNet [25][23], ResNet [26][24], Squeeze-MNet [27][25], and Unet [28][26] networks are typically used in computer vision tasks. However, a CNN needs a large dataset and several layers to understand the global context or relationships between latent features in an image [29][27].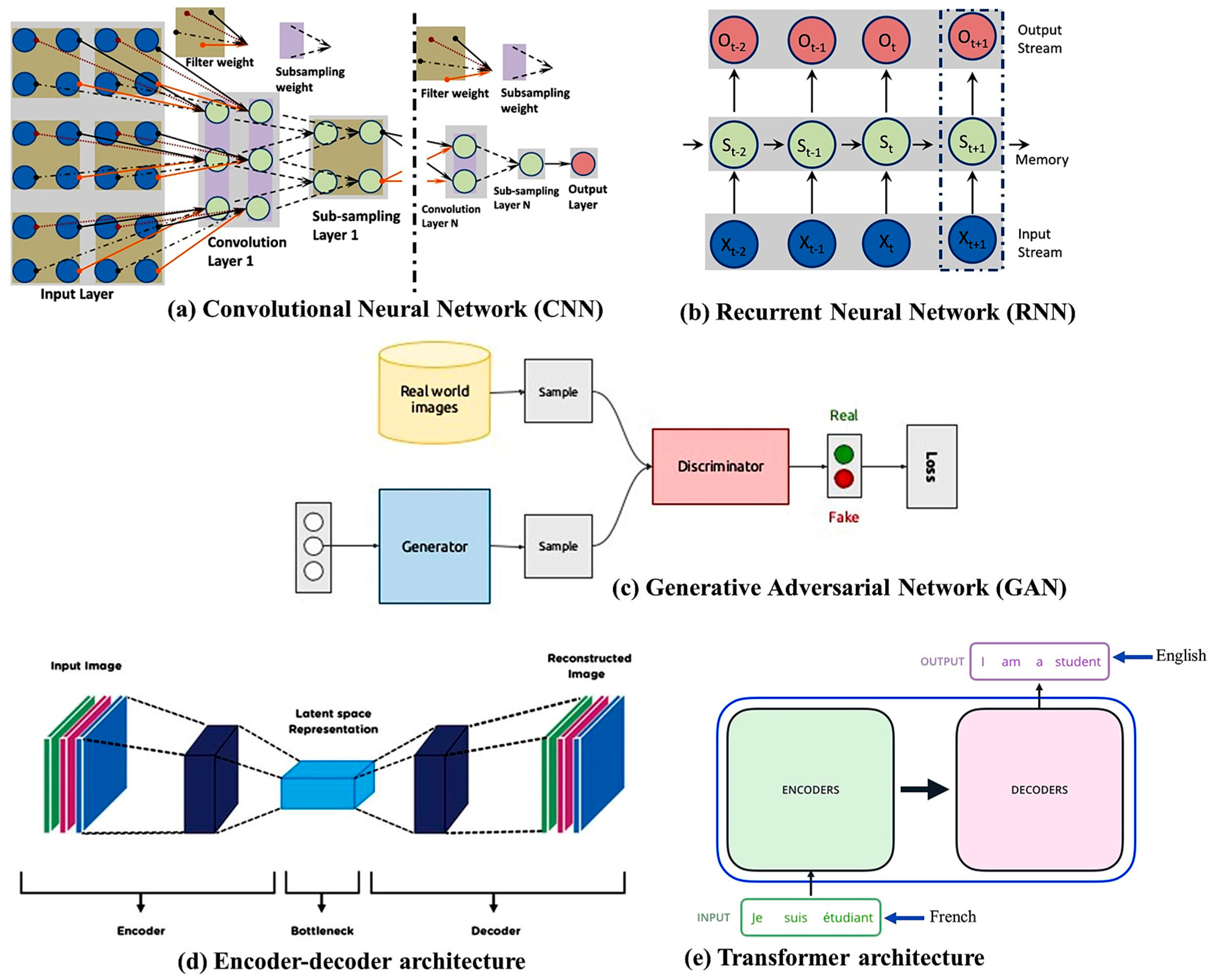
3. DL Tools
DL tools are used to develop models for generating good results. Several popular open-access DL tools used in MRI processing are listed in Table 1. Among them, TensorFlow and PyTorch are widely used.|
Ref. |
Tool Name |
Description |
|---|---|---|
|
Deeplearning4j |
Distributed deep learning library that allows for training models on Java interoperating with the Python environment. |
|
|
Julia |
A flexible and dynamic framework that is more suitable for scientific and numerical computing. |
|
|
Keras |
A Python-based library that is integrated with TensorFlow and used in different ML algorithms. |
|
|
MatConvNet |
A MATLAB toolbox used for image reconstruction, segmentation, and classification by CNN. |
|
|
MS cognitive toolkit |
Describes DNNs as a series of computationally directed graphs, where leaf nodes represent input parameters and other nodes indicate matrix operation. |
|
|
Neural designer |
Data mining tool that was developed by the Artelnics company used in NNs. |
|
|
PyTorch |
Developed by Facebook, works on complex data and is easy to learn. |
|
|
Scikit-image |
Applied for histogram equalization of the input images on various image processing algorithms. |
|
|
Sigpy |
The signal processing package operates on multi-dimensional array plotting and MRI reconstruction. |
|
|
TensorFlow |
Open-source Python framework developed by Google Brain Team that is the most used tool for developing deep learning models. |
|
|
TensorFlow Federated (TFF) |
An open-source framework developed by Google, TFF provides tools for FL. It allows developers to implement federated models and train them across distributed devices. |
|
|
PySyft |
PySyft is a flexible and powerful library for encrypted privacy-preserving ML. It extends PyTorch and TensorFlow to enable the security of FL. |
|
|
Substra |
In 2016, a multi-partner research project developed this FL framework. It concentrates on the medical industry to protect patient privacy and data ownership. It is currently utilized by the pharmaceutical industry for drug discovery. |
4. Network Training Strategies
4.1. Supervised and Unsupervised Learning
Supervised learning is a common technique used in medical image analysis, including the analysis of MRI data. In supervised learning, a machine learning model is trained on a labeled dataset, where each input (in this case, an MRI image) is associated with a corresponding output (typically, a label or annotation). The model learns to map inputs to outputs by identifying patterns and relationships in the training data. Supervised learning in MRI has been applied to a wide range of tasks, including tumor detection and segmentation, disease classification, image registration, and more. It has the potential to significantly enhance the accuracy and efficiency of medical image analysis. However, it also requires large and high-quality labeled datasets and careful validation to ensure its reliability in clinical practice. Unlike supervised learning, where the algorithm is provided with labeled training data (input–output pairs), unsupervised learning [56][54] involves working with unlabeled data. The goal of this learning is to find patterns, structures, or representations in the data without specific guidance regarding the output. Unsupervised learning methods [57,58][55][56] are particularly valuable when dealing with large and complex MRI datasets, as they can reveal hidden structures and patterns within the data without the need for extensive manual labeling. Real-time 3D MRI reconstruction from cine-MRI using unsupervised networks involves leveraging neural networks to reconstruct dynamic 3D MRI volumes from a sequence of 2D images acquired over time (cine-MRI) [59][57]. However, the interpretation of the results obtained from unsupervised learning can be more challenging and often requires domain expertise to make meaningful clinical inferences. These methods are an essential part of the toolkit for researchers and clinicians working with MRI data. Semi-supervised learning [60][58] is a machine learning paradigm that combines elements of both supervised and unsupervised learning. It is particularly useful when you have access to a small amount of labeled data and a large amount of unlabeled data. It is especially valuable in scenarios where acquiring large amounts of labeled data is challenging. This learning can leverage the available labeled data to improve the model performance on tasks such as classification, segmentation, or regression. Semi-supervised learning in MRI analysis offers the advantage of leveraging both labeled and unlabeled data to enhance model performance. By combining the strengths of supervised and unsupervised learning, semi-supervised approaches have the potential to improve the accuracy and robustness of MRI-based diagnostic and analysis tasks. Self-supervised learning [61][59] is an emerging and powerful technique for training machine learning models, especially in scenarios where obtaining labeled data is challenging or expensive. Self-supervised learning is a type of unsupervised learning where the data itself provide supervision for training. This learning in MRI analysis leverages the inherent structure and properties of MRI data to guide the training process, making it a valuable approach for improving the quality of MRI images, enhancing data availability, and addressing various challenges in MRI research and clinical applications. It is an area of active research with the potential to significantly impact the field of medical imaging.4.2. Transfer Learning
Transfer learning (TL) [62][60] is the process of learning a new activity more effectively by transferring the knowledge acquired in one or more source tasks and applying it to the learning of a related target task. The development of methods for knowledge transfer is a step toward making ML as effective as human learning. Using information from the source task, TL aims to enhance learning in the target task. To improve DL network performance, the model complexity is typically increased by raising the architecture’s numbers of layers and nodes. Multiple model parameters must be accurately learned using a large amount of training data. The performance of a model’s reconstruction is typically improved by adding training data. However, because preserving k-space data is not part of the typical clinical flow, it is challenging to obtain patient raw data for training the network. Consequently, the generalizability of a network based on a few samples needs to be improved. Figure 2 shows a diagram of TL, in which the trained model uses the input and reference brain images for learning. After training, it shares the learning knowledge (weights) with a different model to reconstruct an image of a knee.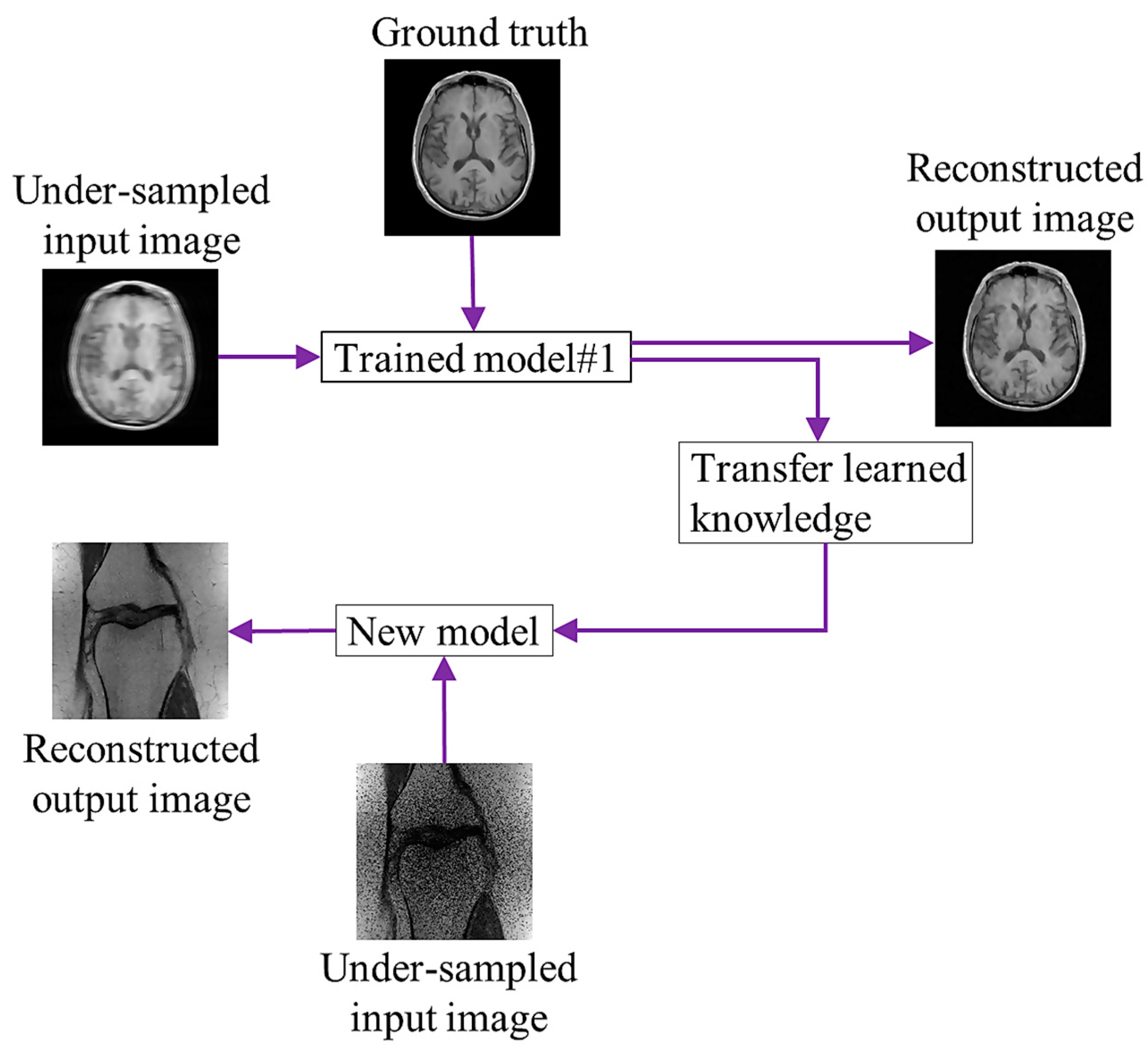
4.3. Federated Learning
Deep networks frequently need large amounts of diversely matched data, which can be labor- and cost-intensive to obtain. Furthermore, retaining patients’ data raises privacy concerns, making it challenging to share the information with other institutions. This problem is addressed by the recently developed FL framework [70][68], which enables the cooperative and distributed training of DL-based techniques. In FL, data are stored locally, and statistical models are trained across segmented data centers or remote devices, e.g., smartphones or hospitals. The training of diverse and possibly large networks poses unexpected problems that call for a fundamental change from conventional methods for large-scale DL, remote optimization, and confidentiality data analysis. To create a global model, a cloud server communicates explicitly with each institution on a regular basis before sharing the data with all the institutions. Each organization uses and maintains its own set of personal information. FL algorithms communicate only about model parameters or update gradients rather than sending actual training data, alleviating privacy concerns. Figure 3 shows communication between global (server side) and local models among several institutions during training. Local models learn from local data and share their weights with the global model.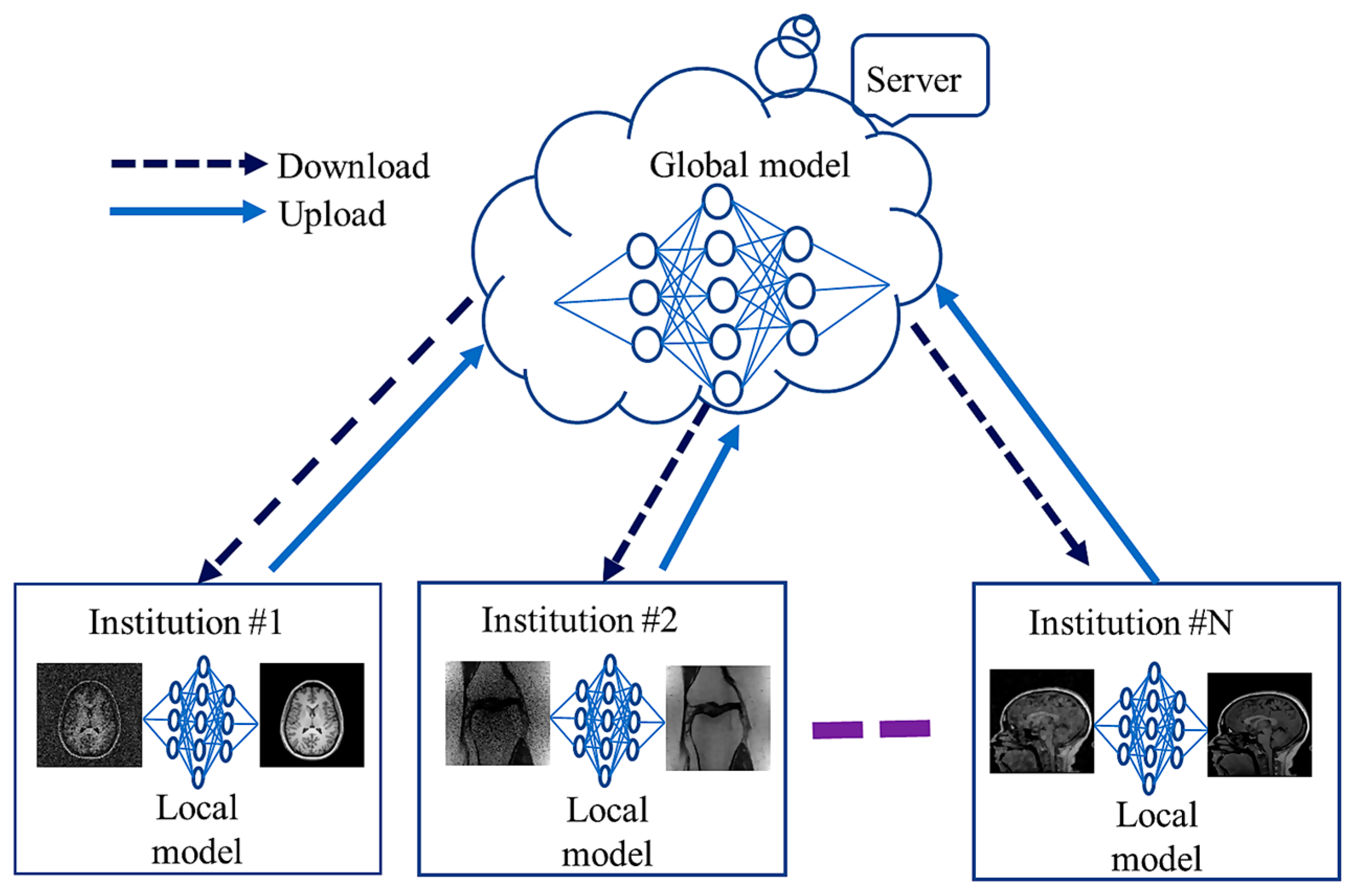
References
- Brown, R.W.; Cheng, Y.-C.N.; Haacke, E.M.; Thompson, M.R.; Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2014; ISBN 9781118633953.
- Cercignani, M.; Dowell, N.G.; Tofts, P.S. Quantitative MRI of the Brain: Principles of Physical Measurement; CRC Press: Boca Raton, FL, USA, 2018; Volume 15, ISBN 9781315363578.
- Muckley, M.J.; Riemenschneider, B.; Radmanesh, A.; Kim, S.; Jeong, G.; Ko, J.; Jun, Y.; Shin, H.; Hwang, D.; Mostapha, M.; et al. Results of the 2020 FastMRI Challenge for Machine Learning MR Image Reconstruction. IEEE Trans. Med. Imaging 2021, 40, 2306–2317.
- Deshmane, A.; Gulani, V.; Griswold, M.A.; Seiberlich, N. Parallel MR Imaging. J. Magn. Reson. Imaging 2012, 36, 55–72.
- Lustig, M.; Donoho, D. Compressed Sensing MRI. Signal Process. Mag. 2008, 25, 72–82.
- Griswold, M.A.; Jakob, P.M.; Heidemann, R.M.; Nittka, M.; Jellus, V.; Wang, J.; Kiefer, B.; Haase, A. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn. Reson. Med. 2002, 47, 1202–1210.
- Pruessmann, K.P.; Weiger, M.; Scheidegger, M.B.; Boesiger, P. SENSE: Sensitivity Encoding for Fast MRI. Magn. Reson. Med. 1999, 42, 952–962.
- Hu, Z.; Zhao, C.; Zhao, X.; Kong, L.; Yang, J.; Wang, X.; Liao, J.; Zhou, Y. Joint Reconstruction Framework of Compressed Sensing and Nonlinear Parallel Imaging for Dynamic Cardiac Magnetic Resonance Imaging. BMC Med. Imaging 2021, 21, 182.
- Islam, R.; Islam, M.S.; Uddin, M.S. Compressed Sensing in Parallel MRI: A Review. Int. J. Image Graph. 2022, 22, 2250038.
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584.
- Zhang, Y.; Gorriz, J.M.; Dong, Z. Deep Learning in Medical Image Analysis. J. Imaging 2021, 7, 74.
- Hossain, M.B.; Kwon, K.-C.; Shinde, R.K.; Imtiaz, S.M.; Kim, N. A Hybrid Residual Attention Convolutional Neural Network for Compressed Sensing Magnetic Resonance Image Reconstruction. Diagnostics 2023, 13, 1306.
- Badža, M.M.; Barjaktarović, M.C. Classification of Brain Tumors from Mri Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999.
- Zhao, C.; Xiang, S.; Wang, Y.; Cai, Z.; Shen, J.; Zhou, S.; Zhao, D.; Su, W.; Guo, S.; Li, S. Context-Aware Network Fusing Transformer and V-Net for Semi-Supervised Segmentation of 3D Left Atrium. Expert Syst. Appl. 2023, 214, 119105.
- Kim, S.; Park, S.; Na, B.; Yoon, S. Spiking-YOLO: Spiking Neural Network for Energy-Efficient Object Detection. Proc. AAAI Conf. Artif. Intell. 2020, 34, 11270–11277.
- Ahishakiye, E.; Van Gijzen, M.B.; Tumwiine, J.; Wario, R.; Obungoloch, J. A Survey on Deep Learning in Medical Image Reconstruction. Intell. Med. 2021, 1, 118–127.
- Montalt-Tordera, J.; Muthurangu, V.; Hauptmann, A.; Steeden, J.A. Machine Learning in Magnetic Resonance Imaging: Image Reconstruction. Phys. Medica 2021, 83, 79–87.
- Zhang, H.M.; Dong, B. A Review on Deep Learning in Medical Image Reconstruction. J. Oper. Res. Soc. China 2020, 8, 311–340.
- He, Z.; Quan, C.; Wang, S.; Zhu, Y.; Zhang, M.; Zhu, Y.; Liu, Q. A Comparative Study of Unsupervised Deep Learning Methods for MRI Reconstruction. Investig. Magn. Reson. Imaging 2020, 24, 179.
- Knoll, F.; Hammernik, K.; Zhang, C.; Moeller, S.; Pock, T.; Sodickson, D.K.; Akcakaya, M. Deep-Learning Methods for Parallel Magnetic Resonance Imaging Reconstruction: A Survey of the Current Approaches, Trends, and Issues. IEEE Signal Process. Mag. 2020, 37, 128–140.
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458.
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A Survey of the Recent Architectures of Deep Convolutional Neural Networks. Artif. Intell. Rev. 2020, 53, 5455–5516.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 25, 145–151.
- Shafiq, M.; Gu, Z. Deep Residual Learning for Image Recognition: A Survey. Appl. Sci. 2022, 12, 8972.
- Shinde, R.K.; Alam, S.; Hossain, B.; Imtiaz, S.; Kim, J. Squeeze-MNet: Precise Skin Cancer Detection Model for Low Computing IoT Devices Using Transfer Learning. Cancers 2023, 14, 12.
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. ISBN 9783319245737.
- Ravi, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.-Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21.
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) Network. Phys. D Nonlinear Phenom. 2020, 404, 132306.
- Ramadevi, R.; Marshiana, D.; Bestley, J.S.; Jamuna, R.D. Recurrent Neural Network (RNN) Analysis for Brain Tumor Classification Using Decision Tree Classifiers. J. Crit. Rev. 2020, 7, 2202–2205.
- Alam, M.S.; Kwon, K.-C.; Md Imtiaz, S.; Hossain, M.B.; Kang, B.-G.; Kim, N. TARNet: An Efficient and Lightweight Trajectory-Based Air-Writing Recognition Model Using a CNN and LSTM Network. Hum. Behav. Emerg. Technol. 2022, 2022, 6063779.
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative Adversarial Networks: An Overview. IEEE Signal Process. Mag. 2018, 35, 53–65.
- Yoon, J.; Jordon, J.; Van Der Schaar, M. Supplementary Materials—RadialGAN: Leveraging Multiple Datasets to Improve Target-Specific Predictive Models Using Generative Adversarial Networks. Int. Conf. Mach. Learn. ICML 2018, 13, 9069–9071.
- Choi, Y.; Choi, M.; Kim, M.; Ha, J.W.; Kim, S.; Choo, J. StarGAN: Unified Generative Adversarial Networks for Multi-Domain Image-to-Image Translation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8789–8797.
- Asadi, A.; Safabakhsh, R. The Encoder-Decoder Framework and Its Applications. In Deep Learning: Concepts and Architectures; Springer: Berlin/Heidelberg, Germany, 2020; pp. 133–167.
- Zhai, J.; Zhang, S.; Chen, J.; He, Q. Autoencoder and Its Various Variants. In Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Miyazaki, Japan, 7–10 October 2018; pp. 415–419.
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. Found. Trends Mach. Learn. 2019, 12, 307–392.
- Patwardhan, N.; Marrone, S.; Sansone, C. Transformers in the Real World: A Survey on NLP Applications. Information 2023, 14, 242.
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 12346, pp. 213–229.
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929.
- Huang, J.; Wu, Y.; Wu, H.; Yang, G. Fast MRI Reconstruction: How Powerful Transformers Are? In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2066–2070.
- Deeplearning4j. Available online: https://deeplearning4j.org/ (accessed on 4 July 2021).
- Julia. Available online: https://julialang.org/ (accessed on 4 July 2021).
- Keras. Available online: https://keras.io/ (accessed on 5 July 2021).
- MatConvNet. Available online: https://www.vlfeat.org/matconvnet/ (accessed on 5 July 2021).
- MS Cognitive Toolkit (CNTK). Available online: https://docs.microsoft.com/en-us/cognitive-toolkit/ (accessed on 5 July 2021).
- Neural Designer. Available online: https://www.neuraldesigner.com/ (accessed on 5 July 2021).
- PyTorch. Available online: https://pytorch.org/ (accessed on 6 July 2021).
- Scikit-Image. Available online: https://scikit-image.org/ (accessed on 6 July 2021).
- Sigpy. Available online: https://sigpy.readthedocs.io/en/latest/ (accessed on 6 July 2021).
- TensorFlow. Available online: https://www.tensorflow.org/ (accessed on 6 July 2021).
- TensorFlow Federated (TFF). Available online: https://www.tensorflow.org/federated (accessed on 15 November 2023).
- PySyft. Available online: https://blog.openmined.org/tag/pysyft/ (accessed on 20 November 2023).
- Substra. Available online: https://www.substra.ai/ (accessed on 10 December 2023).
- Ghahramani, Z. Unsupervised Learning. In Summer School on Machine Learning; Springer: Berlin/Heidelberg, Germany, 2004; pp. 72–112.
- Gong, K.; Han, P.; El Fakhri, G.; Ma, C.; Li, Q. Arterial Spin Labeling MR Image Denoising and Reconstruction Using Unsupervised Deep Learning. NMR Biomed. 2022, 35, e4224.
- Aggarwal, H.K.; Pramanik, A.; John, M.; Jacob, M. ENSURE: A General Approach for Unsupervised Training of Deep Image Reconstruction Algorithms. IEEE Trans. Med. Imaging 2023, 42, 1133–1144.
- Wei, R.; Chen, J.; Liang, B.; Chen, X.; Men, K.; Dai, J. Real-time 3D MRI Reconstruction from Cine-MRI Using Unsupervised Network in MRI-guided Radiotherapy for Liver Cancer. Med. Phys. 2023, 50, 3584–3596.
- Yurt, M.; Dalmaz, O.; Dar, S.; Ozbey, M.; Tinaz, B.; Oguz, K.; Cukur, T. Semi-Supervised Learning of MRI Synthesis without Fully-Sampled Ground Truths. IEEE Trans. Med. Imaging 2022, 41, 3895–3906.
- Hu, C.; Li, C.; Wang, H.; Liu, Q.; Zheng, H.; Wang, S. Self-Supervised Learning for MRI Reconstruction with a Parallel Network Training Framework. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; Springer: Cham, Switzerland, 2021; pp. 382–391.
- Torrey, L.; Shavlik, J. Transfer Learning. In Handbook of Research on Machine Learning Applications and Trends; IGI Global: Hershey, PA, USA, 2010; pp. 242–264.
- Dar, S.U.H.; Özbey, M.; Çatlı, A.B.; Çukur, T. A Transfer-Learning Approach for Accelerated MRI Using Deep Neural Networks. Magn. Reson. Med. 2020, 84, 663–685.
- Arshad, M.; Qureshi, M.; Inam, O.; Omer, H. Transfer Learning in Deep Neural Network Based Under-Sampled MR Image Reconstruction. Magn. Reson. Imaging 2021, 76, 96–107.
- Lv, J.; Li, G.; Tong, X.; Chen, W.; Huang, J.; Wang, C.; Yang, G. Transfer Learning Enhanced Generative Adversarial Networks for Multi-Channel MRI Reconstruction. Comput. Biol. Med. 2021, 134, 104504.
- Yaqub, M.; Jinchao, F.; Ahmed, S.; Arshid, K.; Bilal, M.A.; Akhter, M.P.; Zia, M.S. GAN-TL: Generative Adversarial Networks with Transfer Learning for MRI Reconstruction. Appl. Sci. 2022, 12, 8841.
- Park, S.J.; Ahn, C.-B. Blended-Transfer Learning for Compressed-Sensing Cardiac CINE MRI. Investig. Magn. Reson. Imaging 2021, 25, 10.
- Cheng, C.; Lin, D. MRI Reconstruction Based on Transfer Learning Dynamic Dictionary Algorithm. In Proceedings of the 2023 2nd International Conference on Big Data, Information and Computer Network (BDICN), Xishuangbanna, China, 6–8 January 2023; pp. 1–4.
- Gulamhussene, G.; Rak, M.; Bashkanov, O.; Joeres, F.; Omari, J.; Pech, M.; Hansen, C. Transfer-Learning Is a Key Ingredient to Fast Deep Learning-Based 4D Liver MRI Reconstruction. Sci. Rep. 2023, 13, 11227.
- Yang, Q.; Liu, Y.; Cheng, Y.; Kang, Y.; Chen, T.; Yu, H. Federated Learning; Synthesis Lectures on Artificial Intelligence and Machine Learning Series; Springer: Cham, Switzerland, 2019; Volume 13, pp. 1–207.
- Li, X.; Gu, Y.; Dvornek, N.; Staib, L.H.; Ventola, P.; Duncan, J.S. Multi-Site FMRI Analysis Using Privacy-Preserving Federated Learning and Domain Adaptation: ABIDE Results. Med. Image Anal. 2020, 65, 101765.
- Guo, P.; Wang, P.; Zhou, J.; Jiang, S.; Patel, V.M. Multi-Institutional Collaborations for Improving Deep Learning-Based Magnetic Resonance Image Reconstruction Using Federated Learning. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 2423–2432.
- Feng, C.M.; Yan, Y.; Wang, S.; Xu, Y.; Shao, L.; Fu, H. Specificity-Preserving Federated Learning for MR Image Reconstruction. IEEE Trans. Med. Imaging 2022, 26, 2010–2021.
- Elmas, G.; Dar, S.U.; Korkmaz, Y.; Ceyani, E.; Susam, B.; Ozbey, M.; Avestimehr, S.; Cukur, T. Federated Learning of Generative Image Priors for MRI Reconstruction. IEEE Trans. Med. Imaging 2022, 9, 1996–2009.
- Levac, B.R.; Arvinte, M.; Tamir, J.I. Federated End-to-End Unrolled Models for Magnetic Resonance Image Reconstruction. Bioengineering 2023, 10, 364.
- Feng, C.-M.; Li, B.; Xu, X.; Liu, Y.; Fu, H.; Zuo, W. Learning Federated Visual Prompt in Null Space for MRI Reconstruction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 18–22 June 2023.
- Sandhu, S.S.; Gorji, H.T.; Tavakolian, P.; Tavakolian, K.; Akhbardeh, A. Medical Imaging Applications of Federated Learning. Diagnostics 2023, 13, 3140.
- Li, T.; Sahu, A.K.; Talwalkar, A.; Smith, V. Federated Learning: Challenges, Methods, and Future Directions. IEEE Signal Process. Mag. 2020, 37, 50–60.
