Metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) is a long intergenic non-coding RNA (lncRNA) located on chr11q13. It is overexpressed in several cancers and controls gene expression through chromatin modification, transcriptional regulation, and post-transcriptional regulation. Importantly, MALAT-1 stimulates cell proliferation, migration, and metastasis and serves a vital role in driving the epithelial-to-mesenchymal transition (EMT), subsequently acquiring cancer stem cell-like properties and developing drug resistance. MALAT-1 modulates EMT by interacting with various intracellular signaling pathways, notably the phosphoinositide 3-kinase (PI3K)/Akt and Wnt/β-catenin pathways. It also behaves like a sponge for microRNAs, preventing their interaction with target genes and promoting EMT.
- MALAT-1
- epithelial-to-mesenchymal transition (EMT)
- metastasis
- chemoresistance
1. Introduction
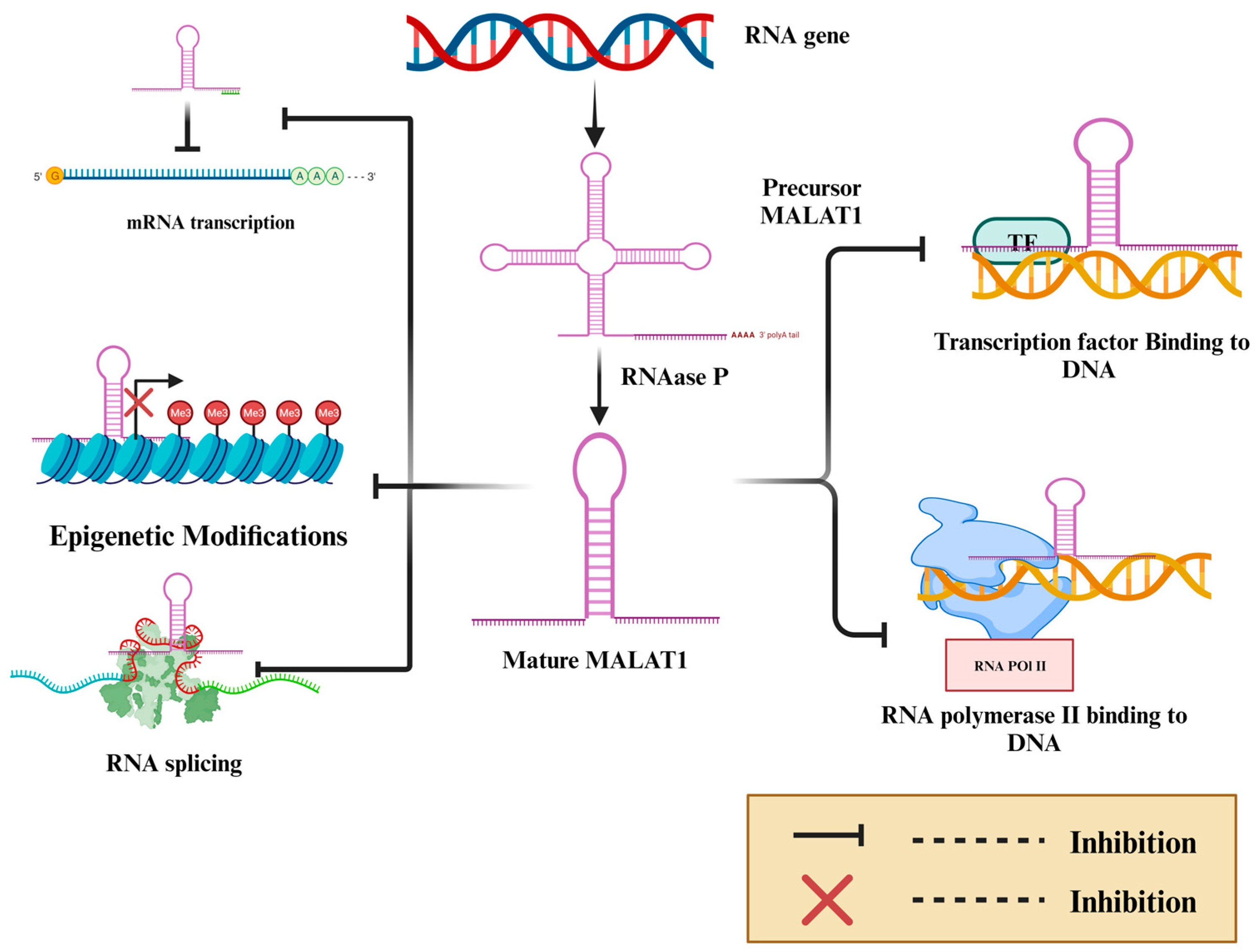
2. MALAT-1 Modulates EMT and Promotes Cancer Metastasis, Stemness, and Chemoresistance
2.1. MALAT-1 Induces Cancer Progression and Metastasis by Modulating EMT
2.2. MALAT-1 Promotes Chemoresistance via Modulating EMT
2.3. MALAT-1 Drives Cancer Cells toward More Stem Cell-like Features by Inducing EMT
References
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041.
- Li, Z.X.; Zhu, Q.N.; Zhang, H.B.; Hu, Y.; Wang, G.; Zhu, Y.S. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 2018, 10, 6757–6768.
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e1435.
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915.e2904.
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851.
- Hartford, C.C.R.; Lal, A. When Long Noncoding Becomes Protein Coding. Mol. Cell Biol. 2020, 40, e00528-19.
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262.
- Huang, J.; Lin, C.; Dong, H.; Piao, Z.; Jin, C.; Han, H.; Jin, D. Targeting MALAT1 induces DNA damage and sensitize non-small cell lung cancer cells to cisplatin by repressing BRCA1. Cancer Chemother. Pharmacol. 2020, 86, 663–672.
- Anbiyaee, O.; Moalemnia, A.; Ghaedrahmati, F.; Shooshtari, M.K.; Khoshnam, S.E.; Kempisty, B.; Halili, S.A.; Farzaneh, M.; Morenikeji, O.B. The functions of long non-coding RNA (lncRNA)-MALAT-1 in the pathogenesis of renal cell carcinoma. BMC Nephrol. 2023, 24, 380.
- Hou, Z.H.; Xu, X.W.; Fu, X.Y.; Zhou, L.D.; Liu, S.P.; Tan, D.M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am. J. Physiol. Cell Physiol. 2020, 318, C649–C663.
- Mekky, R.Y.; Ragab, M.F.; Manie, T.; Attia, A.A.; Youness, R.A. MALAT-1: Immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023, 31, 101653.
- Zhou, Q.; Tang, X.; Tian, X.; Tian, J.; Zhang, Y.; Ma, J.; Xu, H.; Wang, S. LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. J. Cancer 2018, 9, 2436–2442.
- Adewunmi, O.; Shen, Y.; Zhang, X.H.; Rosen, J.M. Targeted Inhibition of lncRNA Malat1 Alters the Tumor Immune Microenvironment in Preclinical Syngeneic Mouse Models of Triple-Negative Breast Cancer. Cancer Immunol. Res. 2023, 11, 1462–1479.
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7.
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014, 35, 2731–2739.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015, 121, 101–108.
- Zheng, H.T.; Shi, D.B.; Wang, Y.W.; Li, X.X.; Xu, Y.; Tripathi, P.; Gu, W.L.; Cai, G.X.; Cai, S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181.
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816.
- Xu, S.; Sui, S.; Zhang, J.; Bai, N.; Shi, Q.; Zhang, G.; Gao, S.; You, Z.; Zhan, C.; Liu, F.; et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 4881–4891.
- Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jiang, W.; Jin, Z.; Guo, Z.; Wang, L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 2014, 32, 2485–2492.
- Dong, Y.; Liang, G.; Yuan, B.; Yang, C.; Gao, R.; Zhou, X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015, 36, 1477–1486.
- Tee, A.E.; Ling, D.; Nelson, C.; Atmadibrata, B.; Dinger, M.E.; Xu, N.; Mizukami, T.; Liu, P.Y.; Liu, B.; Cheung, B.; et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget 2014, 5, 1793–1804.
- Glover, A.R.; Zhao, J.T.; Ip, J.C.; Lee, J.C.; Robinson, B.G.; Gill, A.J.; Soon, P.S.; Sidhu, S.B. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr. Relat. Cancer 2015, 22, 99–109.
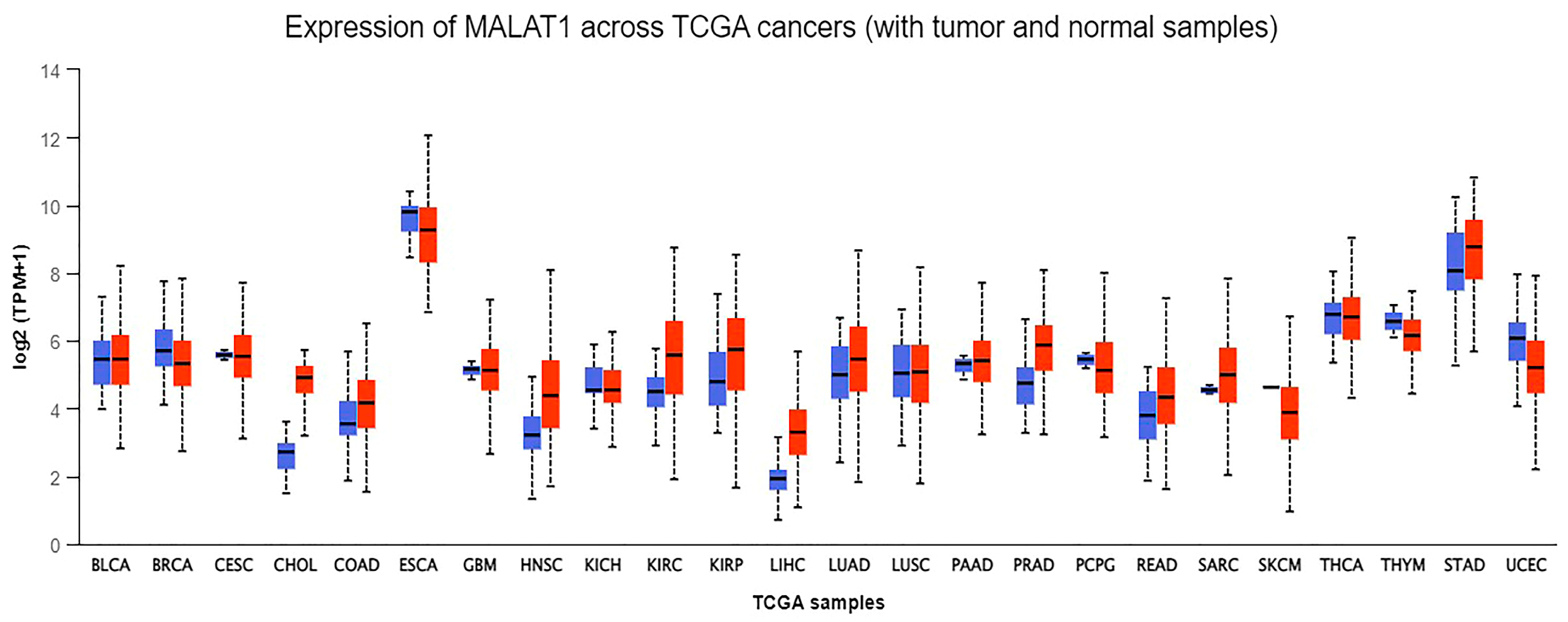
- (UALCAN), The University of Alabama at Birmingham. MALAT-1 Gene Expression across Different Cancers Using Data from The Cancer Genome Atlas (TCGA). Available online: http://ualcan.path.uab.edu/analysis.html (accessed on 20 May 2022).
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27.
- Peng, H.; Wang, J.; Li, J.; Zhao, M.; Huang, S.K.; Gu, Y.Y.; Li, Y.; Sun, X.J.; Yang, L.; Luo, Q.; et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016, 151, 235–242.
- Duan, W.; Du, L.; Jiang, X.; Wang, R.; Yan, S.; Xie, Y.; Yan, K.; Wang, Q.; Wang, L.; Zhang, X.; et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016, 7, 78850–78858.
- Huang, S.K.; Luo, Q.; Peng, H.; Li, J.; Zhao, M.; Wang, J.; Gu, Y.Y.; Li, Y.; Yuan, P.; Zhao, G.H.; et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med. Sci. Monit. 2018, 24, 2476–2488.
- He, B.; Zeng, J.; Chao, W.; Chen, X.; Huang, Y.; Deng, K.; Huang, Z.; Li, J.; Dai, M.; Chen, S.; et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017, 8, 41166–41177.
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087.
- Zhao, Y.; Yu, Y.Q.; You, S.; Zhang, C.M.; Wu, L.; Zhao, W.; Wang, X.M. Long Non-Coding RNA MALAT1 as a Detection and Diagnostic Molecular Marker in Various Human Cancers: A Pooled Analysis Based on 3255 Subjects. Onco Targets Ther. 2020, 13, 5807–5817.
- Xiong, Y.; Wang, J.; Zhu, H.; Liu, L.; Jiang, Y. Chronic oxymatrine treatment induces resistance and epithelialmesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol. Rep. 2018, 39, 967–976.
- Wu, Y.; Sarkissyan, M.; Ogah, O.; Kim, J.; Vadgama, J.V. Expression of MALAT1 Promotes Trastuzumab Resistance in HER2 Overexpressing Breast Cancers. Cancers 2020, 12, 1918.
- Wang, R.; Lu, X.; Yu, R. lncRNA MALAT1 Promotes EMT Process and Cisplatin Resistance of Oral Squamous Cell Carcinoma via PI3K/AKT/m-TOR Signal Pathway. Onco Targets Ther. 2020, 13, 4049–4061.
- Jiao, F.; Hu, H.; Han, T.; Yuan, C.; Wang, L.; Jin, Z.; Guo, Z.; Wang, L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015, 16, 6677–6693.
- Iderzorig, T.; Kellen, J.; Osude, C.; Singh, S.; Woodman, J.A.; Garcia, C.; Puri, N. Comparison of EMT mediated tyrosine kinase inhibitor resistance in NSCLC. Biochem. Biophys. Res. Commun. 2018, 496, 770–777.
- Lee, J.M.; Dedhar, S.; Kalluri, R.; Thompson, E.W. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 2006, 172, 973–981.
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142.
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624.
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274.
- Dong, J.; Hu, Y.; Fan, X.; Wu, X.; Mao, Y.; Hu, B.; Guo, H.; Wen, L.; Tang, F. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 2018, 19, 31.
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454.
- Jechlinger, M.; Grunert, S.; Beug, H. Mechanisms in epithelial plasticity and metastasis: Insights from 3D cultures and expression profiling. J. Mammary Gland. Biol. Neoplasia 2002, 7, 415–432.
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700.
- Niessen, K.; Fu, Y.; Chang, L.; Hoodless, P.A.; McFadden, D.; Karsan, A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008, 182, 315–325.
- Medici, D.; Hay, E.D.; Olsen, B.R. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol. Biol. Cell 2008, 19, 4875–4887.
- Kokudo, T.; Suzuki, Y.; Yoshimatsu, Y.; Yamazaki, T.; Watabe, T.; Miyazono, K. Snail is required for TGFbeta-induced endothelial-mesenchymal transition of embryonic stem cell-derived endothelial cells. J. Cell Sci. 2008, 121, 3317–3324.
- Patel, M.; Eckburg, A.; Gantiwala, S.; Hart, Z.; Dein, J.; Lam, K.; Puri, N. Resistance to Molecularly Targeted Therapies in Melanoma. Cancers 2021, 13, 1115.
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428.
- Garside, V.C.; Chang, A.C.; Karsan, A.; Hoodless, P.A. Co-ordinating Notch, BMP, and TGF-beta signaling during heart valve development. Cell Mol. Life Sci. 2013, 70, 2899–2917.
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland. Biol. Neoplasia 2010, 15, 117–134.
- Wells, A.; Yates, C.; Shepard, C.R. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin. Exp. Metastasis 2008, 25, 621–628.
- Tse, J.C.; Kalluri, R. Mechanisms of metastasis: Epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J. Cell Biochem. 2007, 101, 816–829.
- Sun, R.; Qin, C.; Jiang, B.; Fang, S.; Pan, X.; Peng, L.; Liu, Z.; Li, W.; Li, Y.; Li, G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol. Biosyst. 2016, 12, 952–962.
- Chen, D.; Liu, L.; Wang, K.; Yu, H.; Wang, Y.; Liu, J.; Guo, Y.; Zhang, H. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand. J. Gastroenterol. 2017, 52, 790–796.
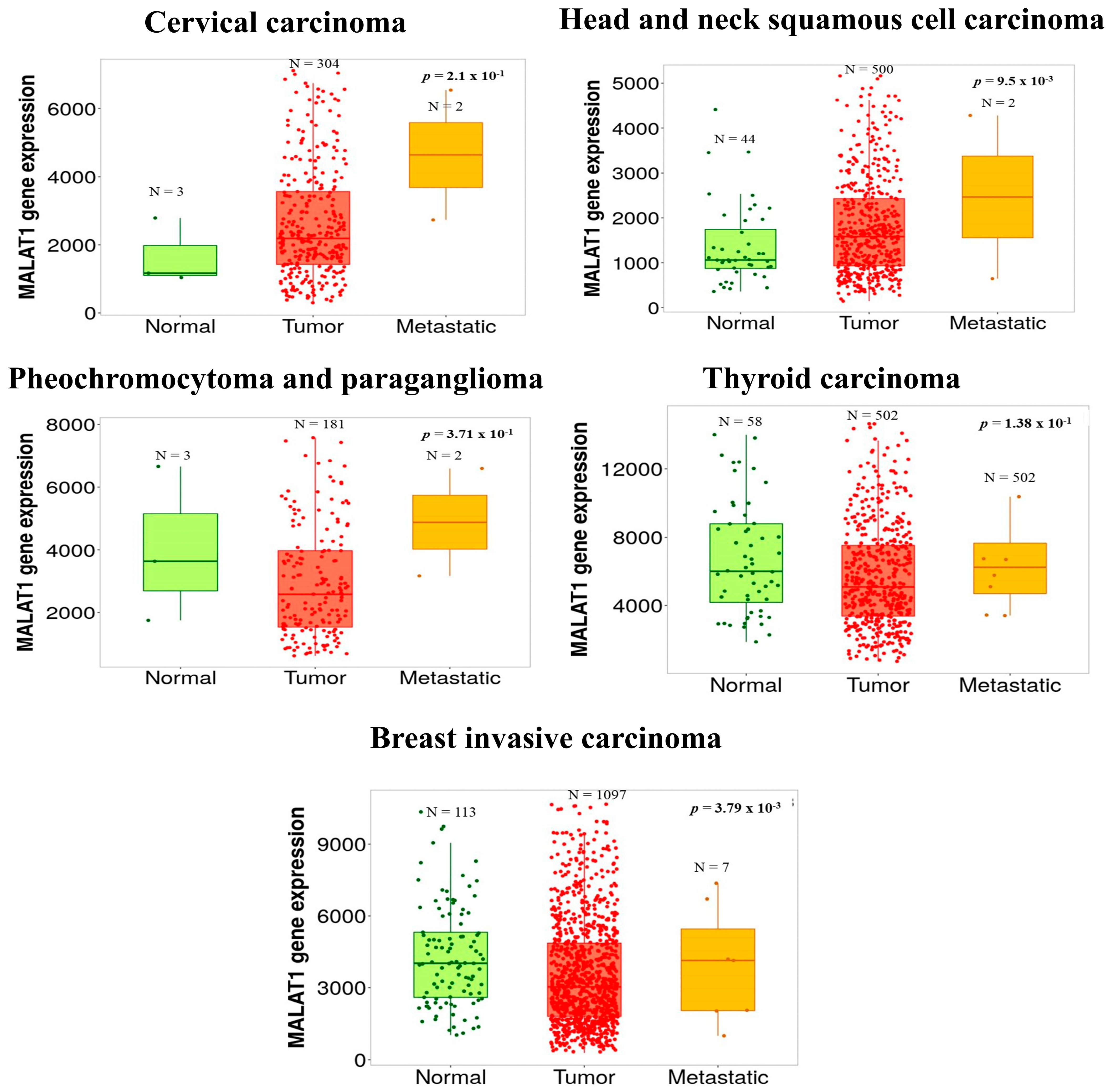
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622.
- Li, H.; Yuan, X.; Yan, D.; Li, D.; Guan, F.; Dong, Y.; Wang, H.; Liu, X.; Yang, B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol. Biochem. 2017, 42, 1192–1201.
- Li, L.J.; Chai, Y.; Guo, X.J.; Chu, S.L.; Zhang, L.S. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2017, 89, 939–948.
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751.
- Zhou, J.; Wang, J.; Zeng, Y.; Zhang, X.; Hu, Q.; Zheng, J.; Chen, B.; Xie, B.; Zhang, W.M. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget 2015, 6, 44332–44345.
- Feng, C.; Zhao, Y.; Li, Y.; Zhang, T.; Ma, Y.; Liu, Y. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch Bronconeumol. 2019, 55, 627–633.
- Zhen, Q.; Liu, J.; Gao, L.; Liu, J.; Wang, R.; Chu, W.; Zhang, Y.; Tan, G.; Zhao, X.; Lv, B. MicroRNA-200a Targets EGFR and c-Met to Inhibit Migration, Invasion, and Gefitinib Resistance in Non-Small Cell Lung Cancer. Cytogenet. Genome Res. 2015, 146, 1–8.
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854.
- Hou, J.; Zhang, G.; Wang, X.; Wang, Y.; Wang, K. Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance. Biomark. Res. 2023, 11, 23.
- Huang, T.; Song, X.; Xu, D.; Tiek, D.; Goenka, A.; Wu, B.; Sastry, N.; Hu, B.; Cheng, S.Y. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics 2020, 10, 8721–8743.
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76.
- Castro-Oropeza, R.; Melendez-Zajgla, J.; Maldonado, V.; Vazquez-Santillan, K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018, 41, 585–603.
- Zeng, L.; Cen, Y.; Chen, J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 2018, 15, 2117–2122.
