The hepatitis E virus (HEV) is a widespread human infection that causes mainly acute infection and can evolve to a chronic manifestation in immunocompromised individuals. In addition to the common strains of hepatitis E virus (HEV-A), known as Paslahepevirus balayani, pathogenic to humans, a genetically highly divergent rat origin hepevirus (RHEV) can cause hepatitis possessing a potential risk of cross-species infection and zoonotic transmission. Rocahepevirus ratti, formerly known as Orthohepevirus C, is a single-stranded RNA virus, recently reassigned to Rocahepevirus genus in the Hepeviridae family, including genotypes C1 and C2.
- hepatitis E
- Rocahepevirus ratti
- Orthohepevirus C
1. Introduction
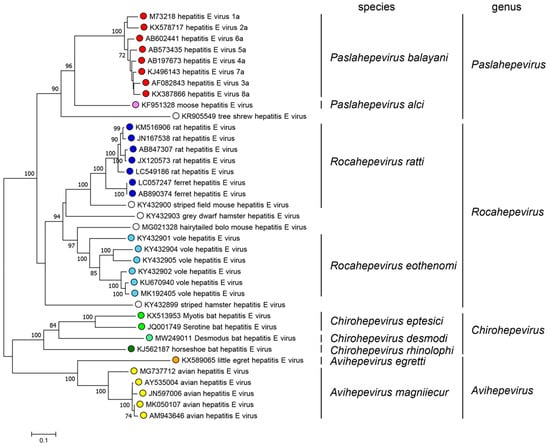
2. Phylogenetic Analysis and Genomic Sequence
The Hepeviridae family is divided into two subfamilies: Orthohepevirinae, divided into four genera: Paslahepevirus (former Orthohepevirus A) the main cause of hepatitis E, which embraces eight different genotypes affecting a variety of species including humans, pigs, wild boar, deer, rabbits, and camels [7]; Rocahepevirus (previous Orthohepevirus C) which infects the Rodentia and Soricomorpha order (rats and ferrets), as well as Carnivora; Chirohepevirus (old Orthohepevirus D) which infects Chiroptera order (bats); and Avihepevirus (previous Orthohepevirus B) which infects and circulates in the avian order (birds); the second subfamily is Parahepevirinae containing one single genus, Piscihepevirus [9][8]. RHEV and HEV-A are two highly divergent viruses, their genomes only share 50–60% genomic identity, differing on their transmission and pathogenicity, with RHEV infections being related to less severe disease outcomes [7]. The HEV virus presents eight main genotypes within the Paslahepevirus genus [4]. Genotypes 1 and 2 transmission is through the consumption of contaminated water by feces, mainly in developing countries. This situation has triggered zoonotic transmission by these genotypes, due to the contamination of the environment with the pathogen from animal feces or urine. As well as the transmission through the ingestion of raw or undercooked meat of the infected animal host, which predominates in genotypes 3 to 8 [5,6][5][6]. Within the RHEV, there are two main genotypes, HEV-C1 and HEV-C2, showing a divergence of 44%. In addition, two extra tentative genotypes (C3 and C4) have been proposed within RHEV species [3]. According to the genomic sequence analysis, the HEV-A virus possesses a single- stranded RNA genome positive-sense approximately 7.2 kb in length. The genome encodes three open reading frames (ORFs) which participate in the viral replication and transcription [9,10][8][9]. ORF1 contains a large polyprotein that is processed into several non- structural proteins, including RNA-dependent RNA polymerase (RdRp), helicase, and protease. ORF2 codifies a protein for the viral capsid, while ORF3 produces a minor protein responsible for the virion assembly and release. The genomic structure of HEV-A resembles other hepeviruses, like the RHEV [11][10]. Recent studies have identified a new open reading frame (ORF4), located within the ORF1 region of the HEV-A genome. This ORF4 encodes a 20 kDa protein with 139–158 amino acids span. Despite its discovery, the role of the ORF4 in the HEV-A or RHEV viral replication and pathogenesis remain unclear [12][11]. Investigating the ORFs of the RHEV virus is crucial to understand the genetic structure of the virus, the pathogenesis, replication, transcription and translation mechanisms, and how they adapt into the host to develop more effective strategies against this pathogen [11][10].3. Epidemiology and Geographical Distribution
3.1. Human Infections Associated with Rocahepevirus ratti
At this moment, up to 21 cases of HEV-C1 infection have been reported in humans through RNA detection in different samples. This data includes sixteen patients in Hong Kong, three patients in Spain, one patient in Central Africa, and one patient in France. RHEV initial detection occurred in rats from Germany and Vietnam [3], but no human cases have been found in Germany [15][12] or Hungary [16][13] after investigation. In Spain, a comprehensive study conducted by Rivero-Juarez et al. revealed the presence of Rocahepevirus ratti infection in three human patients. In China, particularly in Hong Kong, three studies have been carried out to investigate the role of Rocahepevirus ratti. The most recent of these studies was published in 2022 and focused on a cohort of 53 patients from the Microbiology Department of Queen Mary Hospital. The study was conducted between 1 August 2019 and 31 December 2020, and the results indicated a total of eight cases with positive RNA HEV-C1 [17][14]. The study was a continuation of a prior investigation, which analyzed 2201 sera from patients with liver function abnormalities between 1 January 2017 and 31 July 2019 (group 1), and 659 cases in a second group, composed of transplant recipients and patients with solid organ neoplasms, hematologic neoplasms, autoimmune disorders, and other immunosuppressive conditions between 1 January 2019 and 30 June 2019 (group 2). The previous study identified six cases of HEV-C1 RNA positivity in group 1 and one case in group 2, leading to a total of seven cases [18][15]. Notably, the first confirmed human infection caused by RHEV, originating from rats, was identified in Hong Kong in 2017. The study examined a patient who was Anti-HEV IgG positive and had borderline Anti-HEV IgM results. This patient was found to have persistent Hepatitis E after receiving a liver transplant. For this research, a total of 518 solid organ transplant recipients, including kidney, liver, lung, and heart transplants, were evaluated. These recipients exhibited persistent hepatitis, defined as an elevation of alanine aminotransferase (ALT) greater than 1.5 times the upper limit of the reference level for a continuous period of more than six weeks, from 1 January 2014, or the date of transplant (whichever was later) until 31 December 2017 [19][16]. Recent studies have been conducted, revealing also the transmission of RHEV to humans in Europe. A recent study, conducted in Spain, examined 267 cases and evaluated two cohorts: one comprising 169 patients without HEV infection, and the other consisting of 98 patients diagnosed of HEV infection (either HEV RNA or HEV-IgM positive). In the first cohort, two cases of HEV-C1 RNA-positive samples were detected, whereas one case was found in the second cohort (HEV-IgM(+)). Therefore, the first RHEV recorded cases in Europe were these three human infections identified in Spain [4]. A fourth European case was discovered in France thanks to metagenomics testing, revealing the presence of RHEV in liver/blood samples of a male who developed cirrhosis after resolving HBV infection (loss of HBsAg and non detectable DNA).3.2. Animal Infections Associated with Rocahepevirus ratti
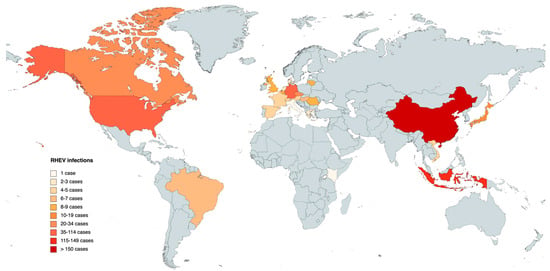
Regarding the discovery and dissemination of RHEV on a global scale, including regions such as Europe, Asia, America, and Africa, the Table 41 provides comprehensive data concerning the prevalence of cases, categorized by continents and individual countries.
| Continent | Asia | n | Europe | n | America | n | Africa | n |
|---|---|---|---|---|---|---|---|---|
| China | 159 | Germany | 42 | USA | 37 | Kenya | 1 | |
| Indonesia | 117 | Czech Republic | 20 | Canada | 21 | |||
| Japan | 14 | Lithuania | 9 | Brazil | 7 | |||
| Vietnam | 5 | Romania | 9 | |||||
| Belgium | 8 | |||||||
| Great Britain | 8 | |||||||
| Hungary | 7 | |||||||
| France | 5 | |||||||
| Country RHEV infections |
Austria | 4 | ||||||
| Spain | 4 | |||||||
| Switzerland | 4 | |||||||
| Denmark | 3 | |||||||
| Greece | 2 | |||||||
| Netherlands | 2 | |||||||
| Italy | 1 | |||||||
| Total infections | 295 | 128 | 65 | 1 |
3.3. Investigation of Rocahepevirus ratti in Wastewater Systems
4. Pathogenesis and Diagnosis
Rocahepevirus ratti, like Paslahepevirus balayani, can cause acute hepatitis, chronic hepatitis, and subclinical infection. HEV-C1 infections are self-limiting and less severe than HEV-A infections, characterized by lower mean peak ALT and bilirubin levels in those with intact immunity. In general, the criteria for inclusion in these investigations to identify RHEV infection include clinical and biological manifestations consistent with acute or chronic hepatitis, as well as an ALT level significantly greater than the upper limit of normal. Infections in immunocompromised individuals, on the other hand, are challenging, with 50% of HEV-C1 infections progressing to persistence, according to research by S. Sridhar et al. [18][15]. This may be due to the fact that immunocompromised people might be more vulnerable to HEV-C1 infections. Not only was the viral pathogenicity examined, but meningoencephalitis was detected as a sequela in an immunocompromised patient after the hepatitis had entirely cured. Therefore, the correlation between RHEV infection and neurological symptoms must be studied. Subclinical HEV-C1 infection, without changes in hepatic function tests or clinical symptoms, is concerning over the safety of blood transfusions because the virus can be transmitted through contaminated blood samples due to the high viral load of HEV-C1 in plasma. RHEV’s viral tropism is oriented towards the liver, with a special affinity for liver cells. Virus replication occurs there, leading to liver damage and the release of liver enzymes into the blood, resulting in acute and chronic hepatitis in humans and animals. Recognizing the pathogenicity is crucial for the development of therapies for the prevention and treatment of hepatitis caused by this virus [18][15]. Ferrets and rats have been proposed as candidate animal models to study RHEV pathogenesis. Ferrets exhibit three patterns of infection: subclinical infection, acute hepatitis, and persistent infection [56][20]. Induced infections of RHEV on immunosuppressed rats have shown the effect of high-dose immunosuppression to induce chronic hepatitis and viral load suppression was observed with ribavirin treatment [57][21]. Molecular biology techniques have facilitated the identification of RHEV RNA in liver, feces, and blood samples obtained from various animal and human species. Such techniques include RT-PCR amplification, Sanger DNA sequencing, and metagenomic tests involving next generation sequencing. The combination of these methods, along with detailed phylogenetic and sequence evaluations, have allowed sequencing of RHEV genomes [4]. The first human RHEV infection was detected in a patient with chronic hepatitis, and this case was validated by RT-PCR and sequencing of plasma, stool, saliva, and liver tissue samples. The most accurate method for detecting Rocahepevirus ratti infection is the RT-PCR method by which viral genomic RNA can be identified. The sensitivity of the method depends on the design and selection of specific primers. The molecular diagnosis of HEV-A and RHEV may be challenging as several assays have shown low sensitivity for endemic genotypes in Europe and the number of genotypes/subtypes described is increasing, therefore, without an evaluation, their sensitivity in this context is unknown. Primers used to detect human HEV may not be able to recognize the RHEV genome in patient samples, limiting the sensitivity of PCR, and only the use of specific primers for RHEV has allowed its detection in different samples. It is important to note that the genetic diversity of HEV-C1 is still unclear, which could also limit the sensitivity of the techniques [19][16].5. Conclusions
An increasing number of cases of animals and humans infected with Rocahepevirus ratti are being detected. Some species of rats have been confirmed as natural reservoirs of RHEV, and they are considered a source of zoonotic infection. It is likely that more cases will be discovered as detection techniques improve. Understanding the virus evolution and diffusion is essential for the development of successful prevention and control measures, treatments, and vaccines. The information obtained from the phylogenetic analysis and genomic sequencing will help researchers track the development of new strains, contributing to epidemiological distribution maps. In addition, this information can also be used to develop diagnostic tests to accurately detect the virus in collected samples and evolve in the discovery of a more precise pathogenesis. These tests are needed to assist healthcare professionals in diagnosing RHEV infections and preventing the spread of the virus. This is a public health concern because it is a new pathogen that can be transmitted to other species and even to humans. Further research is needed to determine the exact route of transmission of the virus to humans, as it remains unknown.References
- National Institute of Diabetes and Digestive and Kidney Diseases. Hepatitis Viral. Available online: https://www.niddk.nih.gov/health-information/informacion-de-la-salud/enfermedades-higado/hepatitis-viral (accessed on 31 October 2023).
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778.
- Reuter, G.; Boros, Á.; Pankovics, P. Review of Hepatitis E Virus in Rats: Evident Risk of Species Orthohepevirus C to Human Zoonotic Infection and Disease. Viruses 2020, 12, 1148.
- Rivero-Juarez, A.; Frias, M.; Perez, A.B.; Pineda, J.A.; Reina, G.; Fuentes-Lopez, A.; Freyre-Carrillo, C.; Ramirez-Arellano, E.; Alados, J.C.; Rivero, A. Orthohepevirus C Infection as an Emerging Cause of Acute Hepatitis in Spain: First Report in Europe. J. Hepatol. 2022, 77, 326–331.
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062.
- Wang, B.; Li, W.; Zhou, J.H.; Li, B.; Zhang, W.; Yang, W.H.; Pan, H.; Wang, L.X.; Bock, C.T.; Shi, Z.L.; et al. Chevrier’s Field Mouse (Apodemus chevrieri) and Père David’s Vole (Eothenomys melanogaster) in China Carry Orthohepeviruses That Form Two Putative Novel Genotypes within the Species Orthohepevirus C. Virol. Sin. 2018, 33, 44–58.
- Wang, B.; Harms, D.; Yang, X.L.; Bock, C.T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis e Virus Variants. Pathogens 2020, 9, 154.
- Seitz, R. Hepatitis e Virus: German Advisory Committee Blood (Arbeitskreis blut), Subgroup “Assessment of Pathogens Transmissible by Blood”. Transfus. Med. Hemother. 2015, 42, 247–265.
- Kenney, S.P.; Meng, X.J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724.
- Wang, B.; Meng, X.J. Structural and Molecular Biology of Hepatitis E Virus. Comput. Struct. Biotechnol. J. 2021, 19, 1907–1916.
- Bai, H.; Ami, Y.; Suzaki, Y.; Doan, Y.H.; Muramatsu, M.; Li, T.C. Open Reading Frame 4 Is Not Essential in the Replication and Infection of Genotype 1 Hepatitis E Virus. Viruses 2023, 15, 784.
- Faber, M.; Wenzel, J.J.; Erl, M.; Stark, K.; Schemmerer, M. No Evidence for Orthohepevirus C in Archived Human Samples in Germany, 2000–2020. Viruses 2022, 14, 742.
- Pankovics, P.; Némethy, O.; Boros, Á.; Pár, G.; Szakály, P.; Reuter, G. Four-Year Long (2014–2017) Clinical and Laboratory Surveillance of Hepatitis E Virus Infections Using Combined Antibody, Molecular, Antigen and Avidity Detection Methods: Increasing Incidence and Chronic HEV Case in Hungary. J. Clin. Virol. 2020, 124, 104284.
- Sridhar, S.; Yip, C.C.Y.; Lo, K.H.Y.; Wu, S.; Situ, J.; Chew, N.F.S.; Leung, K.H.; Chan, H.S.Y.; Wong, S.C.Y.; Leung, A.W.S.; et al. Hepatitis e Virus Species C Infection in Humans, Hong Kong. Clin. Infect. Dis. 2022, 75, 288–296.
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Chew, N.F.S.; Leung, K.H.; Chan, J.F.W.; Zhao, P.S.; Chan, W.M.; Poon, R.W.S.; Tsoi, H.W.; et al. Transmission of Rat Hepatitis E Virus Infection to Humans in Hong Kong: A Clinical and Epidemiological Analysis. Hepatology 2021, 73, 10–22.
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, A.J.X.; Leung, K.H.; Chung, T.W.H.; Chan, J.F.W.; Chan, W.M.; Teng, J.L.L.; et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250.
- Wang, H.; Neyvaldt, J.; Enache, L.; Sikora, P.; Mattsson, A.; Johansson, A.; Lindh, M.; Bergstedt, O.; Norder, H. Variations among Viruses in Influent Water and Effluent Water at a Wastewater Plant over One Year as Assessed by Quantitative PCR and Metagenomics. Appl. Environ. Microbiol. 2020, 86, e02073-20.
- Palombieri, A.; Di Profio, F.; Sarchese, V.; Fruci, P.; Suffredini, E.; Martella, V.; Veneri, C.; Bonanno Ferraro, G.; Mancini, P.; La Rosa, G.; et al. Surveillance for Rat Hepatitis E in Wastewater Networks, Italy. Microbiol. Spectr. 2023, 11, e02675-23.
- Casares-Jimenez, M.; Garcia-Garcia, T.; Suárez-Cárdenas, J.M.; Perez-Jimenez, A.B.; Martín, M.A.; Caballero-Gómez, J.; Michán, C.; Corona-Mata, D.; Risalde, M.A.; Perez-Valero, I.; et al. Correlation of Hepatitis E and Rat Hepatitis E Viruses Urban Wastewater Monitoring and Clinical Cases. Sci. Total Environ. 2024, 908, 168203.
- Li, T.-C.; Yang, T.; Yoshizaki, S.; Ami, Y.; Suzaki, Y.; Ishii, K.; Kishida, N.; Shirakura, M.; Asanuma, H.; Takeda, N.; et al. Ferret Hepatitis E Virus Infection Induces Acute Hepatitis and Persistent Infection in Ferrets. Vet. Microbiol. 2016, 183, 30–36.
- Sridhar, S.; Wu, S.; Situ, J.; Shun, E.H.-K.; Li, Z.; Zhang, A.J.-X.; Hui, K.; Fong, C.H.-Y.; Poon, V.K.-M.; Chew, N.F.-S.; et al. A Small Animal Model of Chronic Hepatitis E Infection Using Immunocompromised Rats. JHEP Rep. 2022, 4, 100546.
