
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marcell Lederer | + 1456 word(s) | 1456 | 2021-05-12 10:43:28 | | | |
| 2 | Rita Xu | -2 word(s) | 1454 | 2021-05-21 04:06:25 | | |
Video Upload Options
MEX3A belongs to the MEX3 (Muscle EXcess) protein family consisting of four members (MEX3A-D) in humans. Characteristic for MEX3 proteins is their domain structure with 2 HNRNPK homology (KH) domains mediating RNA binding and a C-terminal really interesting new gene (RING) domain that harbors E3 ligase function.
1. Introduction
Post-transcriptional control of gene expression influences a variety of cellular processes. In this context, RNA-binding proteins (RBPs) modulate gene expression in various multiprotein complexes via a plethora of different mechanisms, ranging from, e.g., alternative splicing, control of RNA turnover, regulation of mRNA translation, subcellular mRNA localization to post-transcriptional gene editing. In agreement with the versatile role in modulating the fate of genetic information, encompassing coding as well as non-coding RNAs (ncRNAs), deregulation of RBP expression and function severely impacts tumorigenesis. Consistent with cancer presenting a multi-pathway disease and multiple roles of RBPs in modulating gene expression, RBPs influence essentially all cancer hallmarks [1][2][3]. This highlights RBPs as promising therapeutic targets [4][5].
Progress in omic technologies revealed a plethora of canonical as well as novel noncanonical RBPs, unraveling a surprising diversity and plethora of mechanisms guiding the post-transcriptional fate of transcripts. Among 1542 identified RBPs reported in the RBP census [6], only about 350 proteins were categorized within the Gene Ontology molecular function “catalytic activity” [7]. Out of these, 32 RBPs contain validated or putative E3 ligase activity and include all four human members of the MEX3 protein family (Table 1).
Table 1. E3 Ligase domain containing RBPs.
| Gene Symbol | Protein Name | Domain |
|---|---|---|
| AFF4 | AF4/FMR2 family member 4 | UBOX |
| ARIH2 | ariadne RBR E3 ubiquitin protein ligase 2 | RING |
| BARD1 | BRCA1 associated RING domain 1 | RING |
| BRCA1 | breast cancer 1, early onset | RING |
| CNOT4 | CCR4-NOT transcription complex, subunit 4 | RING |
| DZIP3 | DAZ interacting zinc finger protein 3 | RING |
| MEX3A | RNA-binding protein MEX3A | RING |
| MEX3B | RNA-binding protein MEX3B | RING |
| MEX3C | RNA-binding protein MEX3C | RING |
| MEX3D | RNA-binding protein MEX3D | RING |
| MID1 | midline 1 | RING |
| MKRN1 | makorin ring finger protein 1 | RING |
| MKRN2 | makorin ring finger protein 2 | RING |
| MKRN3 | makorin ring finger protein 3 | RING |
| NFX1 | transcriptional repressor NF-X1 isoform 3 | RING |
| PHRF1 | PHD and RING finger domain-containing protein 1 isoform 1 | RING |
| PRPF19 | pre-mRNA processing factor 19 | UBOX |
| RBBP6 | Retinoblastoma-binding protein 6 | RING |
| RC3H1 | ring finger and CCCH-type domains 1 | RING |
| RC3H2 | ring finger and CCCH-type domains 2 | RING |
| RNF113A | RING finger protein 113A | RING |
| RNF113B | RING finger protein 113B | RING |
| RNF17 | RING finger protein 17 isoform 2 | RING |
| SCAF11 | protein SCAF11 | RING |
| TRIM21 | tripartite motif containing 21 | RING |
| TRIM25 | tripartite motif containing 25 | RING |
| TRIM40 | tripartite motif-containing protein 40 isoform a | RING |
| TRIM56 | tripartite motif containing 56 | RING |
| TRIM71 | tripartite motif containing 71, E3 ubiquitin protein ligase | RING |
| UNK | RING finger protein unkempt homolog | RING |
| UNKL | unkempt family zinc finger-like | RING |
| ZNF598 | zinc finger protein 598 | RING |
2. MEX3 Proteins Link the Control of RNA Fate with Protein Ubiquitination
Initially, MEX3 was identified in C. elegans [8], where the protein was implicated in regulating the localized translation of pal-1 mRNA at the posterior of early embryos. The loss of MEX3 leads to ectopic Pal-1 expression resulting in an anterior muscle excess, which set the stage for the name of the protein family. In contrast to its mammalian orthologs, C. elegans MEX3 contains two KH domains mediating RNA binding but lacks a C-terminal RING domain.
The human MEX3 protein family comprises four RNA-binding proteins (RBPs), termed MEX3A-D. In addition to two HNRNPK-homology (KH) domains involved in RNA-binding, human MEX3 proteins are characterized by a C-terminal RING (really interesting new gene) domain (Figure 1a; [9][10]). Recent studies revealed that MEX proteins, especially the so far best-studied paralogs (MEX3A, B and C), modulate oncogenic cell properties, including proliferation, migration, EMT (epithelial-to-mesenchymal-transition) and colony formation, suggesting roles in the regulation of tumor cell self-renewal potential. In agreement, members of the MEX3 protein family are expressed in a variety of cancers where their elevated expression is frequently associated with reduced overall survival probability.
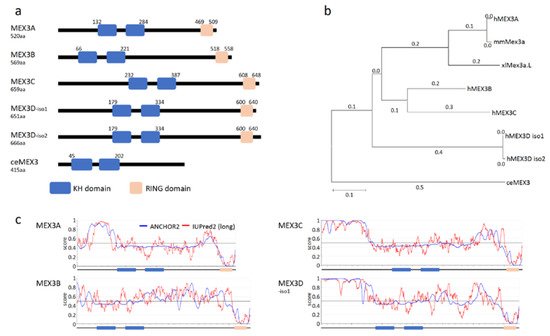
Figure 1. MEX3 family of RNA-binding proteins. (a) Domain structure of human MEX3 proteins and the founder ortholog ceMEX3 of Caenorhabditis elegans with RNA-binding domains (HNRNPK homology domains; KH; blue) and the really interesting new gene (RING domain; orange). Proteins shown: MEX3A (Acc. No. NM001093725.2), MEX3B (Acc. No. NM032246.6), MEX3C (Acc. No. NM016626.5), MEX3D-isoform1 (Acc. No. NM203304.4), MEX3D-isoform2 (Acc. No. NM001174118.2) and MEX3 (Acc. No. NM001381229.2). Annotation and localization of KH di- and RING domain according to www.uniprot.org (accessed on 26 March 2021) [11]. (b) A phylogenetic tree indicating amino acid substitutions of distinct MEX3 paralogs from different species (h: human, mm: Mus musculus, xl: Xenopus laevis and ce: Caenorhabditis elegans). Protein sequences were aligned using Tcoffee before creating a phylogenetic tree with MEGA X. (c) Prediction of intrinsically disordered regions (IUPred2; red) and disordered protein-binding regions (ANCHOR2; blue) using IUPred2A (https://iupred2a.elte.hu; accessed on 30 March 2021).
Mammalian MEX3 proteins are predominantly cytoplasmic but reported to harbor nucleocytoplasmic shuttling activity due to their NES (nuclear export signal) and NLS (nuclear localization signal) motifs. This property suggests that MEX3 proteins may already associate with target transcripts in the nucleus. In the cytoplasm, MEX3A and B localize presumably in an RNA-dependent manner also to P-bodies, whereas MEX3C is evenly distributed in the cytoplasm [9]. P-bodies resemble RNP granules that are due to their composition presumably involved in mRNA turnover, translational control or miRNA-directed regulation of mRNA fate via the RISC complex (for review, see: Ref. [12]). Consistently, MEX3A and B interact in an RNA-dependent manner with Argonaute proteins Ago1 and Ago2. In contrast to MEX3A and B, MEX3C interacts with Ago1 and 2 in an RNA-independent manner [9]. This interaction with components of the RISC complex could delineate a possible mechanism of post-transcriptional gene regulation of this protein family. Additionally, MEX3A enables a noncanonical function of miR-126-5p in the cell nucleus. Ago2-bound miR-126-5p forms a complex with MEX3A on the surface of autophagic vesicles and is transported into the nucleus [13], where miR126-5p prevents caspase-3 dimerization and activation. This supports a dual compartment role, at least of MEX3A.
The canonical domain organization of the four mammalian MEX3 paralogs (54 to 69kDa) is highly conserved (Figure 1b). The overall sequence identity between the paralogs ranges between 37–42% (similarity 45–50%), with a higher degree of identity for the KH1/2 di-domain (78–91%) and the RING domain (68–85%). These similarities suggest similar biochemical functions in RNA binding and E3 ligase activity. Notably, however, the paralogs differ in the position of these functional domains. Whereas the RING domain is positioned at the very C-terminus of the proteins, the N-terminal part, as well as the spacing of the KH1/2-didomain and RING domain (sequence identity 21–39%), varies substantially between paralogs (Figure 1a). These quite divergent sequence regions could contribute to different cellular functions of the paralogs, mediated by a potential association with different factors, e.g., E3 ligase substrates or co-factors modulating MEX3 function, but so far, this hypothesis is not supported by published data. Notably, these regions contain intrinsically disordered stretches (Figure 1c) with disordered protein-binding regions according to IUPred2 and ANCHOR2 prediction [14].
All members of the MEX3 protein family bind RNA irrespective of the organism or cell type. Initial in vitro SELEX studies using the C. elegans MEX-3 KH1/2 di-domain as bait identified an (A/G/U)(G/U)AGN0-8U(U/A/C)UA MEX-3 Recognition Element (MRE) [15]. This was further evaluated by high-resolution protein/RNA co-crystal structures of the individual human MEX3C KH1 (PDB ID 5WWW) and KH2 (PDB ID 5WWX) domains [16]. However, these studies only provide a limited view on the putative role of the MEX3 KH di-domains in RNA binding and shaping of RNA-protein scaffolds for the assembly of larger complexes, as reported for other KH di-domain protein like IGF2BPs [17][18][19][20].
The role of the RING finger domain of MEX3 proteins has mainly been studied in the context of MEX3B and MEX3C. However, all human MEX3 proteins harbor a conserved C-Terminal C3HC4-type RING domain suggesting that all four proteins harbor E3 ligase activity. Crystal structure of MEX3C (PDB ID 5ZI6) in combination with superimposition models with the MDM2–MDMX–UbcH5b–Ub (PDB ID 5MNJ) complex led to the identification of critical residues for auto-ubiquitination as well as E2 ligase interaction [21].
So far, a variety of target mRNAs or proteins ubiquitinated by MEX3 proteins have been proposed. However, there is no comprehensive view on the RNA-binding properties of the protein family since neither CLIP (cross-linking immunoprecipitation) nor RIP (RNA immunopurification) studies have been reported to the best of our knowledge. Likewise, only a few E3 ubiquitin ligase substrates of human MEX3 proteins have been reported. This substantially limits our understanding of how MEX3 proteins may link the control of RNA fate and protein ubiquitination.
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528.
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Xi, T.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 90.
- Mohibi, S.; Chen, X.; Zhang, J. Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer. Pharmacol Ther 2019, 203, 107390.
- Wu, P. Inhibition of RNA-binding proteins with small molecules. Nat. Rev. Chem. 2020, 4, 441–458.
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845.
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338.
- Draper, B.W.; Mello, C.C.; Bowerman, B.; Hardin, J.; Priess, J.R. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 1996, 87, 205–216.
- Buchet-Poyau, K.; Courchet, J.; Le Hir, H.; Seraphin, B.; Scoazec, J.Y.; Duret, L.; Domon-Dell, C.; Freund, J.N.; Billaud, M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007, 35, 1289–1300.
- Pereira, B.; Le Borgne, M.; Chartier, N.T.; Billaud, M.; Almeida, R. MEX-3 proteins: Recent insights on novel post-transcriptional regulators. Trends Biochem. Sci. 2013, 38, 477–479.
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489.
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431.
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourao, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunssen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12.
- Erdos, G.; Dosztanyi, Z. Analyzing Protein Disorder with IUPred2A. Curr. Protoc. Bioinform. 2020, 70, e99.
- Pagano, J.M.; Farley, B.M.; Essien, K.I.; Ryder, S.P. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. USA 2009, 106, 20252–20257.
- Yang, L.; Wang, C.; Li, F.; Zhang, J.; Nayab, A.; Wu, J.; Shi, Y.; Gong, Q. The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity. J. Biol. Chem. 2017, 292, 16221–16234.
- Dagil, R.; Ball, N.J.; Ogrodowicz, R.W.; Hobor, F.; Purkiss, A.G.; Kelly, G.; Martin, S.R.; Taylor, I.A.; Ramos, A. IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties. Nucleic Acids Res. 2019, 47, 4334–4348.
- Chao, J.A.; Patskovsky, Y.; Patel, V.; Levy, M.; Almo, S.C.; Singer, R.H. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010, 24, 148–158.
- Patel, V.L.; Mitra, S.; Harris, R.; Buxbaum, A.R.; Lionnet, T.; Brenowitz, M.; Girvin, M.; Levy, M.; Almo, S.C.; Singer, R.H.; et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012, 26, 43–53.
- Schneider, T.; Hung, L.H.; Aziz, M.; Wilmen, A.; Thaum, S.; Wagner, J.; Janowski, R.; Muller, S.; Schreiner, S.; Friedhoff, P.; et al. Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat. Commun. 2019, 10, 2266.
- Moududee, S.A.; Jiang, Y.; Gilbert, N.; Xie, G.; Xu, Z.; Wu, J.; Gong, Q.; Tang, Y.; Shi, Y. Structural and functional characterization of hMEX-3C Ring finger domain as an E3 ubiquitin ligase. Protein Sci. 2018, 27, 1661–1669.




