
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hongwei Hou | + 2265 word(s) | 2265 | 2021-02-09 13:01:48 | | | |
| 2 | Peter Tang | -17 word(s) | 2248 | 2021-05-19 05:22:33 | | |
Video Upload Options
Plants adapt to environmental changes by regulating their development and growth. As an important interface between plants and their environment, leaf morphogenesis varies between species, populations, or even shows plasticity within individuals. Leaf growth is dependent on many environmental factors, such as light, temperature, and submergence. Phytohormones play key functions in leaf development and can act as molecular regulatory elements in response to environmental signals. In this review, we discuss the current knowledge on the effects of different environmental factors and phytohormone pathways on morphological plasticity and intend to summarize the advances in leaf development. In addition, we detail the molecular mechanisms of heterophylly, the representative of leaf plasticity, providing novel insights into phytohormones and the environmental adaptation in plants.
1. Introduction
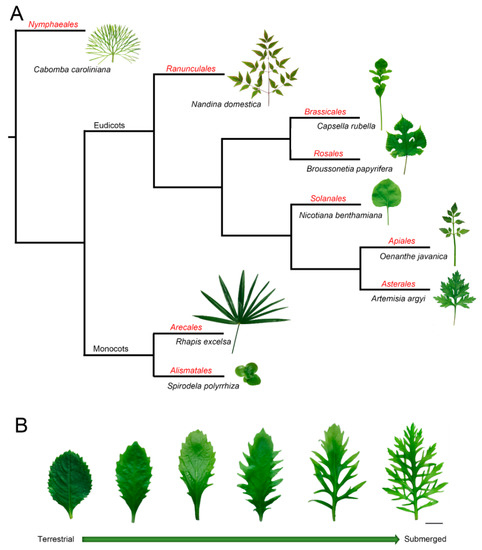
Leaves are key interfaces between plants and their surrounding environment, functioning to capture sunlight, synthesize photosynthate, exchange gasses, sense ambient changes, and regulate their growth under heterogeneous conditions [1][2][3]. In part because of their sessile lifestyle, plants possess efficient systems of morphological plasticity and acclimation to environmental changes. The diversity of leaf shape, vein pattern, stomata, and other parameters not only vary among plants that belong to different species (Figure 1A) but also within a single plant [4][5][6] (Figure 1B). It is well known that the same genotype is capable of developing different phenotypes, which is regarded as the coordination of phenotype, development, and environment [7][8]. For example, heteroblasty was described as the changes in leaf shape during growth development [9], while anisophylly is coupled with asymmetry and leaf phyllotaxis [10]. Some species have even evolved the ability to develop significantly different leaf types under heterogeneous conditions, a phenomenon called heterophylly [11][12][13]. Furthermore, heteroblasty indicates the juvenile-to-adult transition marked by morphological changes, and it emphasizes the developmental stage-related plasticity [14]. However, heterophylly is an extreme morphological plasticity, which is induced by environmental conditions [12][13]. This morphological plasticity provides good models for studying leaf development. However, the mechanisms related to how plants sense environmental changes and develop final leaf forms is still not elucidated.
Given the rapid developments of plant functional genomics, many genes controlling leaf development have been studied, and the regulatory networks underlying these morphological processes have been well characterized [15]. Despite the fact that leaf development is related to genotype, the final shape is adjusted by environmental conditions, such as light, temperature, atmospheric carbon dioxide (CO2) concentrations, and submergence, to adapt to environmental variables [1][16]. The modulation of phytohormone signaling and distributions is a very effective strategy for quick environmental responses. Phytohormones are long-range molecular signals and have key functions in regulating plant growth and leaf development [11][17][18][19][20][21][22][23][24][25][26]. Thus, environmentally induced changes in hormone concentration, distribution, and/or sensitivity can promote coordinated developmental responses [27][28][29][30][31].
2. Environmental Sensing and Adaptation to Light and Temperature
Photosynthesis efficiency depends on the light capture of leaves. As a result, the balance of maximizing light capture and minimizing the harmful impact of high light is a coordinated developmental response. For example, plants prefer to develop broad leaves to maximize light capture, but if the sunlight is too harsh it may lead to overheating and cause harm to the plants [32][33]. In contrast, leaf development also responds to shade (a reduction in the red (660 nm) to far-red (730 nm), R/FR), which is called shade avoidance syndrome (SAS), showing petioles elongation, leaf upward bending, and leaf area decreasing [34][35] (Figure 2A). The upward movement of the leaves allows the plant to elevate the position of the foliage in order to maximize light capture [34][36]. Other aspects are also affected by light, such as leaf complexity, stomata density, and leaf thickness, which increased in the high light conditions [16][37][38][39]. In Rorippa aquatica (Brassicaceae), leaf complexity is dramatically increased in high light conditions [40]. In some other species, such as Nuphar lutea (Nymphaeaceae), Rumex palustris (Polygonaceae), and Hygrophila difformis (Acanthaceae), light change even induced the rearrangement of chloroplasts and altered the photosynthetic biochemistry to adapt the plant to aquatic conditions [41][42][43]. The photoperiod also significantly regulates leaf form. For example, short daylength induced submerged leaves, while long daylength induced terrestrial leaves of P. palustris and Ranunculus aquatilis (Ranunculaceae) [44][45].
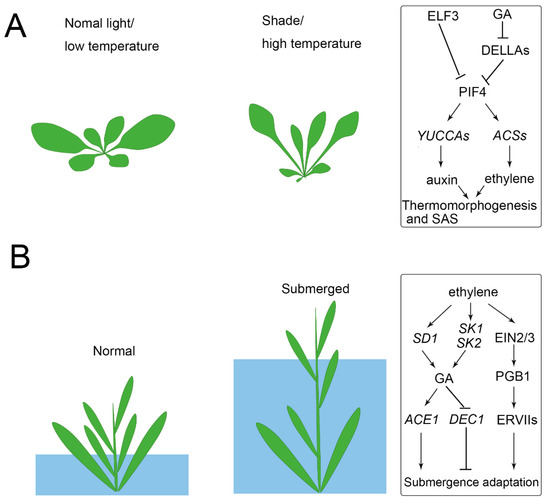
Increasing surrounding temperature affects numerous developmental traits among plants, and the morphological changes that occur in plants in response to temperature changes are called thermomorphogenesis [46][47][48]. In order to adapt to high temperatures, plants developed elongated hypocotyls and petioles, as well as a decrease in leaf thickness and an increasing stomatal density [47][49][50]. These morphological responses are believed to cool plants and reduce the damage caused by sunlight through the upward bending of leaves [46][51][52]. Leaf dissection has for a long-time been thought to correlate with ambient temperature [5]. For example, plants growing in cold climates tend to develop serrated or deep-lobed leaves, while plants growing in warm conditions display shallow-lobed leaves [53][54][55][56][57]. To some degree, leaf dissection was used as an indicator for predicting paleoclimate [5][58]. The change in temperature of a single leaf of R. aquatica affects the epidermal cell size in developing leaves, and hence the morphology of the whole plant is affected [1][59]. In Ludwigia arcuata (Onagraceae), low temperature induced the elongation of epidermal cells and thus lead to the aquatic leaf form [60]. It was recently verified that pectin and cortical microtubules drive morphogenesis in plant epidermal cells [61][62], but how these epidermal changes are regulated by temperature is still unknown.
It was verified that auxin signal functions to connect temperature sensing with growth responses in hypocotyls [63][64]. In Arabidopsis thaliana, temperature changes can be sensed by the inactivation of photoreceptors such as phytochrome B (phyB), whose function in thermoregulation operates via the PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) for high temperature-induced hypocotyl elongation [65][66]. High temperature-activated PIF4 directly upregulates the expression of auxin biosynthesis genes (e.g., YUCCA8, TAA1, and CYB79B2), and as a result, the accumulated auxin induces hypocotyl elongation and leaf hyponasty [67][68]. High temperature also induced PIF4 expression by inactivating EARLY FLOWERING 3 (ELF3) that directly represses PIF4. In high temperature conditions, ELF3 binding to the PIF4 promoter is decreased, and thus PIF4 was activated for thermomorphogenesis [69][70]. Auxin could theoretically induce elongation growth; however, it was recently reported that the phytohormone brassinosteroid (BR) activates elongation growth downstream of auxin to act in themomorphogenesis [71][72].
Temperature and light signals are integrated into the PIF and the relevant genetic network, which controls auxin biosynthesis [67][73]. Photomorphogenesis and shade avoidance responses, including stem/hypocotyl elongation are mediated by PIF4 [74]. The stability of the PIF4 protein is regulated by light, and it is dephosphorylated and stable in the dark, while it is rapidly phosphorylated by phyB-mediated signaling and degradation upon red light irradiation [74]. Interestingly, although phyB and PIF4 antagonistically regulate photomorphogenesis and shade avoidance responses, they cooperatively promote stomatal development in response to high light [39]. Shade also induces the expression of gibberellic acid (GA) biosynthetic enzymes and leads to an accumulation of GA, which then promotes the degradation of DELLAs. It was found that DELLA directly interacts with PIF4 and prevents it from binding to target promoters [75][76]. Besides, the ethylene response also shows short hypocotyls, short roots, and an exaggerated apical hook [77]. PIF4 also promotes ethylene biosynthesis by activating the expression of ethylene biosynthesis genes (e.g., ACS2, 6, 8, and 9) and enhances ethylene signaling by activating the transcription factor ETHYLENE INSENSITIVE 3 (EIN3) [78][79].
Light and temperature are the most critical environmental factors for plant growth, and even a slight change can lead to disasters of plants [80][81]. We mentioned above that PIF4 may be a key element that functions in the light/temperature-dependent morphological plasticity and the crosstalk of phytohormones such as auxin, ethylene, and GA. Future studies based on these gene pathways and phytohormones will not only reveal novel mechanisms on the light and temperature response but will also have implications on crop improvement through use of these plastic strategies.
3. Environmental Sensing and Adaptation to Submergence
Under flooding or submerged conditions, plants find it difficult to obtain enough O2 for respiration. Terrestrial plants, such as A. thaliana and Solanum lycopersicum (tomato), which are intolerant to flooding, find that submerged conditions induce their leaves to turn pale and suppresses their plant growth [73][82]. Deepwater rice survive periodic flooding and consequent oxygen deficiency by activating an internode growth of stems to keep above the water [83] (Figure 2B). In other species such as R. palustris, elongated leaves and decreased thickness helps the plant to obtain a relatively increased gas exchange under submerged conditions [41]. In some aquatic, dimorphic types of plants, their submerged leaves are always thin, narrow, or dissected and contain fewer stomata, while aerial leaves are thick, broad, and entire, and have more stomata [12][40][84][85]. Although narrow or dissected leaves are less efficient at absorbing sunlight than those with wider blades, they can better withstand the destructive force of water flow and more efficiently incorporate CO2 and mineral nutrients than entire leaves [86][87][88].
ABA and ethylene are key regulators of drought and submerge response, separately. ABA was regarded as a stress hormone, which accumulates rapidly in response to drought/dehydration stress and plays a crucial role in stomatal closure, root growth, and the production of protective metabolites [20][89]. ABA levels in unstressed plants are low, but accumulated highly under reduced water potentials by the activation of key synthesis genes 9-cis-epoxycarotenoid dioxygenases (NCEDs) [90]. Upon perception of ABA, the ABA receptor pyrabactin resistance 1 (PYR1)-like protein PYL, regulatory components of the ABA receptor (RCAR) proteins, inhibit the activity of clade A protein phosphatase type 2C (PP2C) phosphatases, thus releasing the subclass III sucrose nonfermenting 1-related kinase 2 (SnRK2s, including SnRK2.2, SnRK2.3, and SnRK2.6) to phosphorylate downstream proteins [91][92]. The arabidopsis protein kinases SnRK2s function as central and positive regulators of the ABA signaling pathway and are involved in stomatal closure, osmotic stress responses, and have an evolutionarily conserved function on plant adaptation to the terrestrial environment [93][94][95].
Aquatic plants, such as rice, have evolved adaptive mechanisms to survive under submergence. When subjected to flooding, rice or deepwater rice accumulates high ethylene, which activates gibberellin biosynthesis gene SEMIDWARF 1 (SD1), promotes GA-dependent elongation, and results in an “escape” strategy to reestablish contact with the air [83]. Recent studies have found that the submergence-induced GA accumulation activates ACCELERATOR OF INTERNODE ELONGATION 1 (ACE1), which confers cells of the intercalary meristematic region with the competence for cell division, leading to internode elongation in the presence of GA. In contrast, high GA repressed DECELERATOR OF INTERNODE ELONGATION 1 (DEC1) suppresses internode elongation, whereas downregulation of DEC1 allows internode elongation [96]. Under submerged conditions, ethylene also induces the expression of two ethylene response factors (ERFs), SNORKEL1 (SK1) and SK2, to trigger remarkable internode elongation via GA [97]. However, the response may vary between species, as GA levels in Rumex acetosa remain unchanged, although ethylene increased during submergence [98]. For the submergence of terrestrial plants, such as A. thaliana, the limited gas diffusion causes passive ethylene accumulation, leading to ETHYLENE INSENSITIVE 2 (EIN2) and EIN3/EIN3-like 1 (EIL1)-dependent signaling and enhanced production of the nitric oxide (NO) scavenger PHYTOGLOBIN 1 (PGB1). The enhanced PGB1 levels lead to NO depletion, enhancing group VII ethylene response factor (ERFVII) stability [99]. The constitutively synthesized ERVIIs (e.g., RELATED TO APETALA 2.12 (RAP2.12), RAP2.2, and RAP2.3) redundantly act as the principal activators of many hypoxia adaptive genes and lead to flooding survival [43].
Phytohormone signals also play key roles in leaf development. For example, the recruitment of leaf founder cells in the shoot apical meristem (SAM) is mediated by the formation of a concentration maxima of auxin [100][101]. Altering the endogenous auxin levels and localization results in leaf simplification in a tomato plant, while downregulating auxin biosynthesis genes (e.g., YUCCA) was reported to inhibit organ initiation in many species such as Arabidopsis, maize, and petunia [102][103][104]. Cytokinin (CK) also plays an important role in SAM maintenance [105][106][107]. Overexpression of the CK biosynthesis genes in tomato leaves leads to the formation of highly compound leaves. However, exogenous application of CK causes minor leaf phenotypes in the tomato [108]. Increasing GA levels in tomatoes result in tall plants with larger and simpler leaves [109]. Interestingly, this GA response is not common, and in some species, GA has the opposite effect of inducing more compound leaves [110][111]. To better understand the relationship of phytohormones and leaf development, in the next section we will discuss the molecular mechanisms of leaf development.
References
- Nakayama, H.; Kimura, S. Leaves may function as temperature sensors in the heterophylly of Rorippa aquatica (Brassicaceae). Plant Signal. Behav. 2015, 10, e1091909.
- Conklin, P.A.; Strable, J.; Li, S.; Scanlon, M.J. On the mechanisms of development in monocot and eudicot leaves. New Phytol. 2019, 221, 706–724.
- Li, G.; Hu, S.; Hou, H.; Kimura, S. Heterophylly: Phenotypic plasticity of leaf shape in aquatic and amphibious plants. Plants 2019, 8, 420.
- Bar, M.; Ori, N. Leaf development and morphogenesis. Development 2014, 141, 4219–4230.
- Chitwood, D.H.; Sinha, N.R. Evolutionary and environmental forces sculpting leaf development. Curr. Biol. 2016, 26, 297–306.
- Tsukaya, H. Leaf shape diversity with an emphasis on leaf contour variation, developmental background, and adaptation. Semin. Cell Dev. Biol. 2018, 79, 48–57.
- Gratani, L. Plant phenotypic plasticity in response to environmental factors. Adv. Bot. 2014, 4, 1–17.
- Sultan, S.E. Developmental plasticity: Re-conceiving the genotype. Interface Focus 2017, 7, 20170009.
- Zotz, G.; Wilhelm, K.; Becker, A. Heteroblasty—A review. Bot. Rev. 2011, 77, 109–151.
- Nakayama, H.; Nakayama, N.; Nakamasu, A.; Sinha, N.; Kimura, S. Toward elucidating the mechanisms that regulate heterophylly. Plant Morphol. 2012, 24, 57–63.
- Nakayama, H.; Sinha, N.R.; Kimura, S. How do plants and phytohormones accomplish heterophylly, leaf phenotypic plasticity, in response to environmental cues. Front. Plant Sci. 2017, 8, 1717.
- Kim, J.; Joo, Y.; Kyung, J.; Jeon, M.; Park, J.Y.; Lee, H.G.; Chung, D.S.; Lee, E.; Lee, I. A molecular basis behind heterophylly in an amphibious plant, Ranunculus trichophyllus. PLoS Genet. 2018, 14, e1007208.
- Li, G.; Hu, S.; Yang, J.; Zhao, X.; Kimura, S.; Schultz, E.A.; Hou, H. Establishment of an Agrobacterium mediated transformation protocol for the detection of cytokinin in the heterophyllous plant Hygrophila difformis (Acanthaceae). Plant Cell Rep. 2020, 39, 737–750.
- Manuela, D.; Xu, M. Juvenile leaves or adult leaves: Determinants for vegetative phase change in higher plants. Int. J. Mol. Sci. 2020, 21, 9753.
- He, D.; Guo, P.; Gugger, P.F.; Guo, Y.; Liu, X.; Chen, J. Investigating the molecular basis for heterophylly in the aquatic plant Potamogeton octandrus (Potamogetonaceae) with comparative transcriptomics. PeerJ 2018, 6, e4448.
- Tsukaya, H. Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 2005, 49, 547–555.
- Sanz, L.; Albertos, P.; Mateos, I.; Sanchez-Vicente, I.; Lechon, T.; Fernandez-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868.
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359.
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344.
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118.
- Peres, A.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the sixth class of phytohormones: A molecular view from the discovery to hormonal interactions in plant development and stress adaptation. Int. J. Mol. Sci. 2019, 20, 331.
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565.
- Su, Y.; Liu, Y.; Zhang, X. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625.
- Vanstraelen, M.; Benkova, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487.
- Zhu, Z.; Lee, B. Friends or foes: New insights in jasmonate and ethylene co-actions. Plant Cell Physiol. 2015, 56, 414–420.
- Nguyen, T.N.; Tuan, P.A.; Mukherjee, S.; Son, S.; Ayele, B.T. Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat. J. Exp. Bot. 2018, 69, 4065–4082.
- Kohli, A.; Sreenivasulu, N.; Lakshmanan, P.; Kumar, P.P. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013, 32, 945–957.
- Fahad, S.; Hussain, S.; Bano, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.A.; Khan, F.; Chen, Y.; Wu, C.; et al. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: Consequences for changing environment. Environ. Sci. Pollut. Res. Int. 2015, 22, 4907–4921.
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marin-de la Rosa, N.; Sauer, M.; Wenig, M.; Villaecija-Aguilar, J.A.; et al. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276.
- Phukan, U.J.; Mishra, S.; Shukla, R.K. Waterlogging and submergence stress: Affects and acclimation. Crit. Rev. Biotechnol. 2016, 36, 956–966.
- Brunetti, C.; Sebastiani, F.; Tattini, M. Review: ABA, flavonols, and the evolvability of land plants. Plant Sci. 2019, 280, 448–454.
- Ort, D. When there is too much light. Plant Physiol. 2001, 125, 29–32.
- Weraduwage, S.M.; Chen, J.; Anozie, F.C.; Morales, A.; Weise, S.E.; Sharkey, T.D. The relationship between leaf area growth and biomass accumulation in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 167.
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427.
- Pantazopoulou, C.K.; Bongers, F.J.; Kupers, J.; Reinen, E.; Das, D.; Evers, J.B.; Anten, N.; Pierik, R. Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, 7450–7455.
- Carabelli, M.; Possenti, M.; Sessa, G.; Ciolfi, A.; Sassi, M.; Morelli, G.; Ruberti, I. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007, 21, 1863–1868.
- Lake, J.; Quick, W.; Beerling, D.; Woodward, I. Plant development—Signals from mature to new leaves. Nature 2001, 411, 154.
- Yano, S.; Terashima, I. Developmental process of sun and shade leaves in Chenopodium album L. Plant Cell Environ. 2004, 27, 781–793.
- Casson, S.A.; Franklin, K.A.; Gray, J.E.; Grierson, C.S.; Whitelam, G.C.; Hetherington, A.M. phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 2009, 19, 229–234.
- Nakayama, H.; Nakayama, N.; Seiki, S.; Kojima, M.; Sakakibara, H.; Sinha, N.; Kimura, S. Regulation of the KNOX-GA gene module induces heterophyllic alteration in North American lake cress. Plant Cell 2014, 26, 4733–4748.
- Mommer, L.; Pons, T.L.; Wolters-Arts, M.; Venema, J.H.; Visser, E.J. Submergence-induced morphological, anatomical, and biochemical responses in a terrestrial species affect gas diffusion resistance and photosynthetic performance. Plant Physiol. 2005, 139, 497–508.
- Horiguchi, G.; Nemoto, K.; Yokoyama, T.; Hirotsu, N. Photosynthetic acclimation of terrestrial and submerged leaves in the amphibious plant Hygrophila difformis. AoB Plants 2019, 11, plz009.
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020.
- Wallenstein, A.; Albert, L. Plant morphology: Its control in Proserpinaca by photoperiod, temperature, and gibberellic acid. Science 1963, 140, 998–1000.
- COOK, C. On the determination of leaf form in Ranunculus aquatilis. New Phytol. 1969, 68, 469–480.
- Ibanez, C.; Poeschl, Y.; Peterson, T.; Bellstadt, J.; Denk, K.; Gogol-Doring, A.; Quint, M.; Delker, C. Ambient temperature and genotype differentially affect developmental and phenotypic plasticity in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 114.
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 15190.
- Casal, J.J.; Balasubramanian, S. Thermomorphogenesis. Annu Rev. Plant Biol. 2019, 70, 321–346.
- Erwin, J.; Heins, R.; Karlsson, M. Thermomorphogenesis in Lilium longiflorum. Am. J. Bot. 1989, 76, 47–52.
- Fritz, M.A.; Rosa, S.; Sicard, A. Mechanisms underlying the environmentally induced plasticity of leaf morphology. Front. Genet. 2018, 9, 478.
- Crawford, A.J.; McLachlan, D.H.; Hetherington, A.M.; Franklin, K.A. High temperature exposure increases plant cooling capacity. Curr. Biol. 2012, 22, R396–R397.
- Bridge, L.J.; Franklin, K.A.; Homer, M.E. Impact of plant shoot architecture on leaf cooling: A coupled heat and mass transfer model. J. R. Soc. Interface 2013, 10, 20130326.
- Bailey, I.W.; Sinnott, E.W. The climatic distribution of certain types of angiosperm leaves. Am. J. Bot. 1916, 3, 24–39.
- Webb, L. Environmental relationships of the structural types of Australian rain forest vegetation. Ecology 1968, 49, 296.
- Royer, D.L.; Meyerson, L.A.; Robertson, K.M.; Adams, J.M. Phenotypic plasticity of leaf shape along a temperature gradient in Acer rubrum. PLoS ONE 2009, 4, e7653.
- Peppe, D.J.; Royer, D.L.; Cariglino, B.; Oliver, S.Y.; Newman, S.; Leight, E.; Enikolopov, G.; Fernandez-Burgos, M.; Herrera, F.; Adams, J.M.; et al. Sensitivity of leaf size and shape to climate: Global patterns and paleoclimatic applications. New Phytol. 2011, 190, 724–739.
- Chitwood, D.H.; Rundell, S.M.; Li, D.Y.; Woodford, Q.L.; Yu, T.T.; Lopez, J.R.; Greenblatt, D.; Kang, J.; Londo, J.P. Climate and developmental plasticity: Interannual variability in grapevine leaf morphology. Plant Physiol. 2016, 170, 1480–1491.
- Little, S.A.; Kembel, S.W.; Wilf, P. Paleotemperature proxies from leaf fossils reinterpreted in light of evolutionary history. PLoS ONE 2010, 5, e15161.
- Amano, R.; Nakayama, H.; Morohoshi, Y.; Kawakatsu, Y.; Ferjani, A.; Kimura, S. A decrease in ambient temperature induces post-mitotic enlargement of palisade cells in North American Lake Cress. PLoS ONE 2015, 10, e0141247.
- Sato, M.; Tsutsumi, M.; Ohtsubo, A.; Nishii, K.; Kuwabara, A.; Nagata, T. Temperature-dependent changes of cell shape during heterophyllous leaf formation in Ludwigia arcuata (Onagraceae). Planta 2008, 228, 27–36.
- Haas, K.; Wightman, R.; Meyerowitz, E.; Peaucelle, A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science 2020, 367, 1003–1007.
- Li, Y.; Deng, M.; Liu, H.; Li, Y.; Chen, Y.; Jia, M.; Xue, H.; Shao, J.; Zhao, J.; Qi, Y.; et al. ABNORMAL SHOOT 6 interacts with KATANIN 1 and SHADE AVOIDANCE 4 to promote cortical microtubule severing and ordering in Arabidopsis. J. Integr. Plant Biol. 2020.
- Delker, C.; Sonntag, L.; James, G.V.; Janitza, P.; Ibanez, C.; Ziermann, H.; Peterson, T.; Denk, K.; Mull, S.; Ziegler, J.; et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014, 9, 1983–1989.
- Bellstaedt, J.; Trenner, J.; Lippmann, R.; Poeschl, Y.; Zhang, X.; Friml, J.; Quint, M.; Delker, C. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 2019, 180, 757–766.
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009, 19, 408–413.
- Stavang, J.A.; Gallego-Bartolome, J.; Gomez, M.D.; Yoshida, S.; Asami, T.; Olsen, J.E.; Garcia-Martinez, J.L.; Alabadi, D.; Blazquez, M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009, 60, 589–601.
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235.
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594.
- Box, M.S.; Huang, B.E.; Domijan, M.; Jaeger, K.E.; Khattak, A.K.; Yoo, S.J.; Sedivy, E.L.; Jones, D.M.; Hearn, T.J.; Webb, A.A.R.; et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 2015, 25, 194–199.
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976.
- Ibanez, C.; Delker, C.; Martinez, C.; Burstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e303.
- Martinez, C.; Espinosa-Ruiz, A.; de Lucas, M.; Bernardo-Garcia, S.; Franco-Zorrilla, J.M.; Prat, S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018, 37, e99552.
- Mizutani, M.; Kanaoka, M.M. Environmental sensing and morphological plasticity in plants. Semin. Cell Dev. Biol. 2018, 83, 69–77.
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323.
- de Lucas, M.; Daviere, J.M.; Rodriguez-Falcon, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blazquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484.
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479.
- Guzman, P.; Ecker, J. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523.
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636.
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787.
- Franklin, K.A. Light and temperature signal crosstalk in plant development. Curr. Opin. Plant Biol. 2009, 12, 63–68.
- Kostaki, K.I.; Coupel-Ledru, A.; Bonnell, V.C.; Gustavsson, M.; Sun, P.; McLaughlin, F.J.; Fraser, D.P.; McLachlan, D.H.; Hetherington, A.M.; Dodd, A.N.; et al. Guard cells integrate light and temperature signals to control stomatal aperture. Plant Physiol. 2020, 182, 1404–1419.
- Tang, H.; Bi, H.; Liu, B.; Lou, S.; Song, Y.; Tong, S.; Chen, N.; Jiang, Y.; Liu, J.; Liu, H. WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021, 229, 106–125.
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186.
- Robionek, A.; Banas, K.; Chmara, R.; Szmeja, J. The avoidance strategy of environmental constraints by an aquatic plant Potamogeton alpinus in running waters. Ecol. Evol. 2015, 5, 3327–3337.
- Iida, S.; Ikeda, M.; Amano, M.; Sakayama, H.; Kadono, Y.; Kosuge, K. Loss of heterophylly in aquatic plants: Not ABA-mediated stress but exogenous ABA treatment induces stomatal leaves in Potamogeton perfoliatus. J. Plant Res. 2016, 129, 853–862.
- Wanke, D. The ABA-mediated switch between submersed and emersed life-styles in aquatic macrophytes. J. Plant Res. 2011, 124, 467–475.
- van Veen, H.; Sasidharan, R. Shape shifting by amphibious plants in dynamic hydrological niches. New Phytol. 2021, 229, 79–84.
- Mitsui, Y.; Nomura, N.; Isagi, Y.; Tobe, H.; Setoguchi, H. Ecological barriers to gene flow between riparian and forest species of Ainsliaea (Asteraceae). Evolution 2011, 65, 335–349.
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Sig. Behav. 2007, 2, 135–138.
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970.
- Ma-Lauer, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068.
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071.
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363.
- Fujii, H.; Zhu, J. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385.
- Shinozawa, A.; Otake, R.; Takezawa, D.; Umezawa, T.; Komatsu, K.; Tanaka, K.; Amagai, A.; Ishikawa, S.; Hara, Y.; Kamisugi, Y.; et al. SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2019, 2, 30.
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114.
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030.
- He, C.; Morgan, P.; Drew, M. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiol. 1996, 112, 463–472.
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379.
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260.
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911.
- Zhao, Y.; Christensen, S.; Fankhauser, C.; Cashman, J.; Cohen, J.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309.
- Phillips, K.; Skirpan, A.; Liu, X.; Masterson, A.; Slewinski, T.; Hudson, C.; Barazesh, S.; Cohen, J.; Malcomber, S.; McSteen, P. vanishing tassel2 Encodes a Grass-Specific Tryptophan Aminotransferase Required for Vegetative and Reproductive Development in Maize. Plant Cell 2011, 23, 550–566.
- Shwartz, I.; Levy, M.; Ori, N.; Bar, M. Hormones in tomato leaf development. Dev. Biol. 2016, 419, 132–142.
- Gordon, S.; Chickarmane, V.; Ohno, C.; Meyerowitz, E. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2009, 106, 16529–16534.
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655.
- Shani, E.; Ben-Gera, H.; Shleizer-Burko, S.; Burko, Y.; Weiss, D.; Ori, N. Cytokinin regulates compound leaf development in tomato. Plant Cell 2010, 22, 3206–3217.
- Fleishon, S.; Shani, E.; Ori, N.; Weiss, D. Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 2011, 190, 609–617.
- Jasinski, S.; Tattersall, A.; Piazza, P.; Hay, A.; Martinez-Garcia, J.F.; Schmitz, G.; Theres, K.; McCormick, S.; Tsiantis, M. PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 2008, 56, 603–612.
- DeMason, D.A.; Chetty, V.J. Interactions between GA, auxin, and UNI expression controlling shoot ontogeny, leaf morphogenesis, and auxin response in Pisum sativum (Fabaceae): Or how the uni-tac mutant is rescued. Am. J. Bot. 2011, 98, 775–791.
- Li, G.; Hu, S.; Yang, J.; Schultz, E.A.; Clarke, K.; Hou, H. Water-Wisteria as an ideal plant to study heterophylly in higher aquatic plants. Plant Cell Rep. 2017, 36, 1225–1236.





