Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Eric A. Weaver | + 2121 word(s) | 2121 | 2021-04-29 06:20:50 | | | |
| 2 | Catherine Yang | Meta information modification | 2121 | 2021-05-12 05:42:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Weaver, E.A.; Bullard, B. Influenza Hemagglutinin Vaccines. Encyclopedia. Available online: https://encyclopedia.pub/entry/9545 (accessed on 03 March 2026).
Weaver EA, Bullard B. Influenza Hemagglutinin Vaccines. Encyclopedia. Available at: https://encyclopedia.pub/entry/9545. Accessed March 03, 2026.
Weaver, Eric A., Brianna Bullard. "Influenza Hemagglutinin Vaccines" Encyclopedia, https://encyclopedia.pub/entry/9545 (accessed March 03, 2026).
Weaver, E.A., & Bullard, B. (2021, May 12). Influenza Hemagglutinin Vaccines. In Encyclopedia. https://encyclopedia.pub/entry/9545
Weaver, Eric A. and Brianna Bullard. "Influenza Hemagglutinin Vaccines." Encyclopedia. Web. 12 May, 2021.
Copy Citation
Hemagglutinin (HA) is the predominant antigenic protein of influenza viruses and antibodies directed at HA are correlated with protection against influenza virus infection
universal vaccine
stalk
headless
1. Hemagglutinin Structure and Function
Hemagglutinin is the most abundant protein on the surface of influenza and functions in viral entry through receptor binding and membrane fusion [1]. HA is also the predominant antigenic protein and therefore shows the highest rates of adaptive evolution out of all the influenza proteins [2]. Although there are high levels of protein sequence diversity between the subtypes, the HA protein maintains required elements, such as the cleavage site, secretory signal, fusion domain, transmembrane domain, and cytoplasmic tail as well as common protein structural motifs [3].
HA is a glycoprotein which assembles as a homotrimer on the surface of the virion (Figure 1) [1]. Each monomer starts initially as a single polypeptide precursor (HA0) which is later cleaved into HA1 and HA2 subunits by host proteases. This cleavage is essential for maturation of the virus to an infectious virion [4]. The HA2 subunit is composed mostly of the stalk region of HA and the C-terminus which has a transmembrane domain with a cytoplasmic tail that anchors the HA protein to the envelope of the influenza virion. The HA1 subunit contains the signal peptide at the N-terminus and the globular head domain. This globular head domain contains the receptor binding site which binds sialic acid on the surface of the host cell and facilitates viral entry [5]. Upon internalization of the virion, acidification of the endosome induces a conformational change of HA, which exposes the N-terminus fusion peptide of the HA2 subunit. The fusion peptide then facilitates membrane fusion and release of the viral RNA into the cytoplasm of the host cell [4][6].
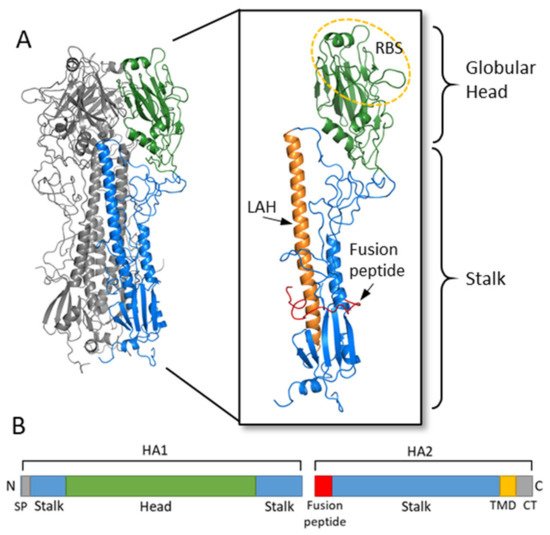
Figure 1. Hemagglutinin structure and functional regions. (A) The HA trimer of an H3N2 virus was downloaded from the Protein Data Bank (PDB: 1HGF; A/X-31) and visualized with PyMOL. Two monomers are colored in grey while the third monomer shows the head region in green and the stalk region in blue. An enlarged view of the HA monomer is further colored to show the fusion peptide in red, the long alpha helix (LAH) in orange, and the receptor binding site (RBS) on the head circled in yellow. (B) A linear schematic of the HA molecular is shown below. The head domain (green) is on the HA1 subunit, while the stalk domain (blue) spans the C- and N-terminus of HA1 along with most of HA2. At the N-terminus of HA1 is the signal peptide (SP) while at the N-terminus of HA2 is the fusion peptide. The transmembrane domain (TMD) and cytoplasmic tail (CT) are at the C-terminus of HA2.
The globular head of influenza also contains the antigenic sites determined for both H1 and H3 [5]. Neutralizing antibodies for influenza are typically directed against these highly antigenic sites on the globular head and interfere with HA binding to sialic acid [5]. The HA protein of influenza virus has the ability to agglutinate red blood cells. Anti-influenza virus antibodies which bind to HA and inhibit the hemagglutination activity of HA are used as a surrogate measure of determining neutralizing antibody titers (HI titers) [7]. Human serological studies have demonstrated that HI titers of at least 1:40 correlate with protection from influenza infection [7][8]. Antibodies directed against the stalk domain of HA have different mechanisms of action, as discussed below, and cannot be measured using an HI assay.
2. Consensus-Based Strategies
2.1. General Principles of Consensus-Based Stratgies
Unlike stalk-directed strategies, consensus-based approaches target the full-length HA protein and aim to design an HA protein which is broadly representative of the diverse HA population. These HA genes are computationally designed and therefore are synthetic in nature. Although the central concept behind consensus-based strategies is similar, many different approaches have been developed (Figure 2). Importantly, unlike stalk-directed strategies, consensus-based approaches typically induce HI antibodies directed against the head region of HA, which is generally accepted as a correlate of protection against influenza infection in humans [9][10]. Consensus-based approaches are typically developed for a single subtype of influenza, with the intention of expanding the approach to multiple subtypes and combining the vaccine constructs into a single multivalent vaccine.
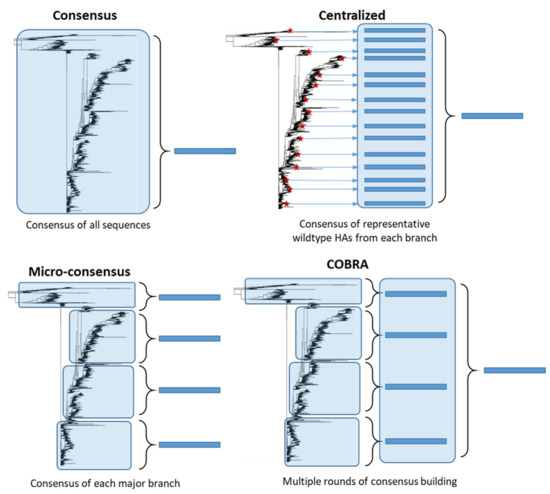
Figure 2. Consensus-based antigen design strategies. Consensus strategies are designed by aligning the target HA protein sequence population and determining the most common amino acid at each position. ‘Consensus’ immunogens are designed using all the HA protein sequences from the phylogenetic tree. ‘Micro-consensus’ immunogens are created by taking a consensus of each major branch of the phylogenetic tree and delivering the immunogens as a cocktail. ‘Centralized’ immunogens aim to reduce sampling bias by taking a representative wildtype HA (red stars) from each branch of the phylogenetic tree and designing a consensus of those HA protein sequences. ‘COBRA’ immunogens also aim to reduce sampling bias and use multiple rounds of consensus building.
2.2. Design of Consensus-Based Vaccines
2.2.1. Consensus Hemagglutinins
Consensus HA genes are constructed by taking the most common amino acid at each position of the HA after aligning a protein sequence population. A consensus strategy has been explored by multiple groups to target highly pathogenic avian influenza (HPAI) H5N1 virus [11][12][13][14], which poses a substantial pandemic threat. In addition, H5N1 has high viral diversity, with multiple clades and subclades [15]. Two immunizations of mice with a consensus H5 gene expressed in a DNA plasmid induced cross-reactive antibodies against multiple different H5 viruses from two clades [11]. Vaccination conferred complete protection from morbidity and mortality after challenge with two H5 clades but only partial protection after challenge with more distantly related H5 viruses. Additionally, a consensus H5 protein expressed in VLPs completely protected chickens from lethal challenge with two H5 viruses from separate clades [13]. Importantly, these consensus immunogens are designed using all H5 protein sequences in the database, which can lead to geographical sampling/sequence bias and result in a consensus HA that might not accurately reflect the HA diversity of the population.
2.2.2. Micro-Consensus Hemagglutinin
While other consensus designs develop a single consensus HA per subtype, Elliott et al. (2018) explored the efficacy of four micro-consensus immunogens to improve cross-reactivity to the H3 subtype [16]. These four micro-consensus genes were expressed in a DNA plasmid and administered twice as cocktail. Vaccination of mice with this cocktail induced strong antibody responses against 8 strains circulating between 1968 and 2014 as measured by ELISA and induced significant HA-specific cellular immunity. Vaccination also protected mice from lethal challenge with two H3N2 strains, with 5–10% weight loss. Importantly, this study finds that a cocktail of consensus immunogens might improve cross-reactivity of highly diverse HA populations, such as the H3 subtype.
2.2.3. Centralized Hemagglutinins
Another consensus-based strategy aims to develop a synthetic HA which localizes to the central node of the phylogenetic tree, thereby minimizing genetic and antigenic differences of unmatched strains. An important limitation to consensus strategy described above is the threat of sampling bias leading to generation of a synthetic HA gene which does not accurately represent the diversity of the population and is biased towards an overrepresented geographical location. Weaver et al. (2011) overcame this by developing a centralized H1 gene using selected representative wildtype HA protein sequences from each major branch of the phylogenetic tree and designing a consensus of those sequences [17]. In this way, each branch is equally represented to prevent sampling/sequencing bias and, as a result, this synthetic HA gene localizes to the central node of the tree. This centralized H1 gene was expressed in a replication-defective adenovirus vector and vaccination of mice resulted in better cross-protection against three H1 strains as compared to mismatched wild type HAs or traditional influenza vaccines after lethal influenza challenge [17]. This strategy was then applied to develop H3 and H5 centralized genes which also demonstrated improved homosubtypic cross-protection [18]. Importantly, a combination of H1, H2, H3, and H5 centralized genes into a single multivalent vaccine did not diminish the increased cross-reactive protection [19]. A single immunization at the high dose of this multivalent formulation demonstrated complete protection from morbidity and mortality after lethal challenge with three H1 strains, three H5 strains, and one H3 strain with partial cross-protection against another two H3 strains. This validates the approach of designing multiple subtype-specific broadly reactive HA immunogens which can then be combined into one multivalent vaccine without reduction in cross-reactivity or interference between immunogens.
2.2.4. COBRA Hemagglutinins
Like the centralized HA design, another antigen design method called computationally optimized broadly cross-reactive antigen (COBRA) aims to minimize sampling/sequencing bias in the target population. The COBRA strategy achieves this through multiple rounds of consensus generation. A COBRA vaccine strategy has been developed and tested for H1 [20][21][22][23][24], H2 [25], H3 [26][27], H5 [28][29][30][31][32][33][34][35], and swine H1 [36]. These COBRA immunogens have primarily been expressed using a VLP platform but have also been explored using ferritin nanoparticles [23] and recombinant live influenza virus [22]. This strategy was first explored as a H5 vaccine targeting only clade 2 and was tested in mice, ferrets [28][29], chickens [34], and non-human primates (NHP) [30]. Vaccination of NHP induced cross-reactive HI antibody titers against multiple clade 2 viruses and against a clade 1 and 7 influenza virus. This strategy was then expanded to target the entirety of the H5 diversity by using a cocktail of three COBRA immunogens designed for human clade 2, human and avian clade 2, and all H5 clades [31]. Vaccination of mice with two doses of this cocktail induced protective HI titers to twenty-five H5 viruses from eleven H5 clades/subclades, however cross-protection was only evaluated after immunization with individual COBRA immunogens and was not evaluated after vaccination with the cocktail.
The COBRA strategy was also evaluated for protection against seasonal and pandemic H1 strains [21]. Nine different COBRA H1 immunogens were designed and four immunogens were selected for further investigation based on their increased cross-reactivate HI titer and protection. These immunogens were evaluated in sequential prime/boost immunizations strategies or as a heterologous cocktail. Results found that the broadest HI titers were induced by a combination of COBRA immunogens designed using both seasonal and pandemic H1 strains, with a trivalent cocktail demonstrating protective HI titers to ten of the fifteen H1 strains [21]. This increased HI cross-reactivity extended to a pre-immune ferret model [20]. However, cross-protection was only evaluated against a single pandemic strain.
A similar strategy was used for the H3 subtype, in which seventeen CORBA immunogens were designed with only four of these immunogens showing promising HI titers in mice [26]. The four immunogenic H3 COBRA immunogens were further explored in a ferret model [27]. Although vaccination of naïve mice showed promising cross-reactivity, vaccination of naïve ferrets showed low HI titers and narrow cross-reactivity, with the most cross-reactive immunogen showing protective HI titers to only six of the thirteen H3 strains. In contrast, ferrets that were pre-immune to a historical H3 strain prior to vaccination showed increased cross-reactive HI titers against the entire panel of 13 viruses after vaccination with COBRA immunogens as compared to vaccination with a wildtype HA. This led the authors theorize that, although there was limited efficacy in naïve ferret, the COBRA immunogens were more effective than wildtype HA at recalling broad cross-reactive memory B cells from previous influenza infection. Therefore, while this H3 COBRA vaccine might boost cross-reactive immunity in pre-immune adults, more research needs to be performed to examine potential efficacy in naïve populations, such as children. In addition, although the COBRA strategy has been explored for multiple influenza subtypes individually, COBRA has not yet been explored as a multivalent formulation targeting multiple subtypes.
2.3. Challenges Facing Consensus-Based Approaches
Consensus-based approaches target the full-length HA protein and therefore typically induce antibodies directed against the variable head region. For this reason, these vaccines are subtype-specific and do not show the cross-reactivity between subtypes of the same phylogenetic group, as is observed in stalk-directed strategies discussed above. However, antibodies induced by these consensus-based approaches have improved cross-reactivity within a subtype and often demonstrate HI activity which is a correlate of protection against influenza infection in humans [9][10]. Therefore, what these approaches lack in breadth of immunity between subtypes, they make up for in robustness of protection within subtypes.
Consensus-based influenza vaccines based on the HA protein are a more recent concept than the stalk-directed strategies and therefore have not progressed to human clinical trials yet. In addition, unlike stalk-based strategies, little work has been performed to evaluate the potential for escape mutants from pre-existing immunity induced by consensus vaccines. The subtype-specific immunity induced by consensus vaccines will necessitate a multivalent cocktail of HA immunogens to protect against the multiple subtypes currently circulating in humans. Further work to demonstrate that a multivalent vaccine containing many immunodominant antigenic sites from different subtypes does not reduce efficacy through interference. However, the centralized HA approach has shown that vaccination of mice with a quadrivalent vaccine showed no reduction in efficacy, indicating that this a promising strategy to induce a robust protective immune response against multiple subtypes relevant to human health.
References
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013.
- Bhatt, S.; Holmes, E.C.; Pybus, O.G. The Genomic Rate of Molecular Adaptation of the Human Influenza A Virus. Mol. Biol. Evol. 2011, 28, 2443–2451.
- Nobusawa, E.; Aoyama, T.; Kato, H.; Suzuki, Y.; Tateno, Y.; Nakajima, K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 1991, 182, 475–485.
- Steinhauer, D.A. Role of Hemagglutinin Cleavage for the Pathogenicity of Influenza Virus. Virology 1999, 258, 1–20.
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4).
- Worch, R. Structural biology of the influenza virus fusion peptide. Acta Biochim. Pol. 2014, 61.
- Coudeville, L.; Bailleux, F.; Riche, B.; Megas, F.; Andre, P.; Ecochard, R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: Development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010, 10, 18.
- Black, S.; Nicolay, U.; Vesikari, T.; Knuf, M.; Del Giudice, G.; Della Cioppa, G.; Tsai, T.; Clemens, R.; Rappuoli, R. Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr. Infect Dis. J. 2011, 30, 1081–1085.
- Nath Neerukonda, S.; Vassell, R.; Weiss, C.D. Neutralizing Antibodies Targeting the Conserved Stem Region of Influenza Hemagglutinin. Vaccines 2020, 8, 382.
- Ellebedy, A.H.; Webby, R.J. Influenza vaccines. Vaccine 2009, 27, D65–D68.
- Chen, M.-W.; Cheng, T.-J.R.; Huang, Y.; Jan, J.-T.; Ma, S.-H.; Yu, A.L.; Wong, C.-H.; Ho, D.D. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. USA 2008, 105, 13538–13543.
- Laddy, D.J.; Yan, J.; Kutzler, M.; Kobasa, D.; Kobinger, G.P.; Khan, A.S.; Greenhouse, J.; Sardesai, N.Y.; Draghia-Akli, R.; Weiner, D.B. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS ONE 2008, 3, e2517.
- Wu, P.; Lu, J.; Zhang, X.; Mei, M.; Feng, L.; Peng, D.; Hou, J.; Kang, S.-M.; Liu, X.; Tang, Y. Single Dose of Consensus Hemagglutinin-Based Virus-Like Particles Vaccine Protects Chickens against Divergent H5 Subtype Influenza Viruses. Front. Immunol. 2017, 8.
- Laddy, D.J.; Yan, J.; Corbitt, N.; Kobinger, G.P.; Weiner, D.B. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 2007, 25, 2984–2989.
- Ducatez, M.F.; Bahl, J.; Griffin, Y.; Stigger-Rosser, E.; Franks, J.; Barman, S.; Vijaykrishna, D.; Webb, A.; Guan, Y.; Webster, R.G.; et al. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc. Natl. Acad. Sci. USA 2011, 108, 349–354.
- Elliott, S.T.C.; Keaton, A.A.; Chu, J.D.; Reed, C.C.; Garman, B.; Patel, A.; Yan, J.; Broderick, K.E.; Weiner, D.B. A Synthetic Micro-Consensus DNA Vaccine Generates Comprehensive Influenza A H3N2 Immunity and Protects Mice Against Lethal Challenge by Multiple H3N2 Viruses. Hum. Gene Ther. 2018, 29, 1044–1055.
- Weaver, E.A.; Rubrum, A.M.; Webby, R.J.; Barry, M.A. Protection against Divergent Influenza H1N1 Virus by a Centralized Influenza Hemagglutinin. PLoS ONE 2011, 6, e18314.
- Webby, R.J.; Weaver, E.A. Centralized Consensus Hemagglutinin Genes Induce Protective Immunity against H1, H3 and H5 Influenza Viruses. PLoS ONE 2015, 10, e0140702.
- Lingel, A.; Bullard, B.L.; Weaver, E.A. Efficacy of an Adenoviral Vectored Multivalent Centralized Influenza Vaccine. Sci. Rep. 2017, 7, 14912.
- Carter, D.M.; Darby, C.A.; Johnson, S.K.; Carlock, M.A.; Kirchenbaum, G.A.; Allen, J.D.; Vogel, T.U.; Delagrave, S.; DiNapoli, J.; Kleanthous, H.; et al. Elicitation of Protective Antibodies against a Broad Panel of H1N1 Viruses in Ferrets Preimmune to Historical H1N1 Influenza Viruses. J. Virol. 2017, 91.
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J. Virol. 2016, 90, 4720–4734.
- Sautto, G.A.; Kirchenbaum, G.A.; Ecker, J.W.; Bebin-Blackwell, A.G.; Pierce, S.R.; Ross, T.M. Elicitation of Broadly Protective Antibodies following Infection with Influenza Viruses Expressing H1N1 Computationally Optimized Broadly Reactive Hemagglutinin Antigens. ImmunoHorizons 2018, 2, 226–237.
- Sautto, G.A.; Kirchenbaum, G.A.; Abreu, R.B.; Ecker, J.W.; Pierce, S.R.; Kleanthous, H.; Ross, T.M. A Computationally Optimized Broadly Reactive Antigen Subtype-Specific Influenza Vaccine Strategy Elicits Unique Potent Broadly Neutralizing Antibodies against Hemagglutinin. J. Immunol. 2020, 204, 375–385.
- Darricarrère, N.; Pougatcheva, S.; Duan, X.; Rudicell, R.S.; Chou, T.H.; DiNapoli, J.; Ross, T.M.; Alefantis, T.; Vogel, T.U.; Kleanthous, H.; et al. Development of a Pan-H1 Influenza Vaccine. J. Virol. 2018, 92.
- Reneer, Z.B.; Jamieson, P.J.; Skarlupka, A.L.; Huang, Y.; Ross, T.M. Computationally Optimized Broadly Reactive H2 HA Influenza Vaccines Elicited Broadly Cross-Reactive Antibodies and Protected Mice from Viral Challenges. J. Virol. 2020, 95, e01526-20.
- Wong, T.M.; Allen, J.D.; Bebin-Blackwell, A.G.; Carter, D.M.; Alefantis, T.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Computationally Optimized Broadly Reactive Hemagglutinin Elicits Hemagglutination Inhibition Antibodies against a Panel of H3N2 Influenza Virus Cocirculating Variants. J. Virol. 2017, 91.
- Allen, J.D.; Jang, H.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Elicitation of Protective Antibodies against 20 Years of Future H3N2 Cocirculating Influenza Virus Variants in Ferrets Preimmune to Historical H3N2 Influenza Viruses. J. Virol. 2019, 93, e00946-18.
- Giles, B.M.; Ross, T.M. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 2011, 29, 3043–3054.
- Giles, B.M.; Bissel, S.J.; Dealmeida, D.R.; Wiley, C.A.; Ross, T.M. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin. Vaccine Immunol. Cvi. 2012, 19, 128–139.
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schultz-Cherry, S.; Wiley, C.A.; Ross, T.M. A Computationally Optimized Hemagglutinin Virus-Like Particle Vaccine Elicits Broadly Reactive Antibodies that Protect Nonhuman Primates from H5N1 Infection. J. Infect. Dis. 2012, 205, 1562–1570.
- Crevar, C.J.; Carter, D.M.; Lee, K.Y.; Ross, T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccin Immunother. 2015, 11, 572–583.
- Allen, J.D.; Owino, S.O.; Carter, D.M.; Crevar, C.J.; Reese, V.A.; Fox, C.B.; Coler, R.N.; Reed, S.G.; Baldwin, S.L.; Ross, T.M. Broadened immunity and protective responses with emulsion-adjuvanted H5 COBRA-VLP vaccines. Vaccine 2017, 35, 5209–5216.
- Bar-Peled, Y.; Huang, J.; Nuñez, I.A.; Pierce, S.R.; Ecker, J.W.; Ross, T.M.; Mousa, J.J. Structural and antigenic characterization of a computationally-optimized H5 hemagglutinin influenza vaccine. Vaccine 2019, 37, 6022–6029.
- Ross, T.M.; DiNapoli, J.; Giel-Moloney, M.; Bloom, C.E.; Bertran, K.; Balzli, C.; Strugnell, T.; Mariana, S.E.S.; Mebatsion, T.; Bublot, M.; et al. A computationally designed H5 antigen shows immunological breadth of coverage and protects against drifting avian strains. Vaccine 2019, 37, 2369–2376.
- Nuñez, I.A.; Ross, T.M. Human COBRA 2 vaccine contains two major epitopes that are responsible for eliciting neutralizing antibody responses against heterologous clades of viruses. Vaccine 2020, 38, 830–839.
- Skarlupka, A.L.; Owino, S.O.; Suzuki-Williams, L.P.; Crevar, C.J.; Carter, D.M.; Ross, T.M. Computationally optimized broadly reactive vaccine based upon swine H1N1 influenza hemagglutinin sequences protects against both swine and human isolated viruses. Hum. Vaccin Immunother. 2019, 15, 2013–2029.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
12 May 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





