
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stefania Stella | + 2603 word(s) | 2603 | 2021-01-14 07:02:55 | | | |
| 2 | Dean Liu | -811 word(s) | 1792 | 2021-05-07 05:23:54 | | | | |
| 3 | Dean Liu | Meta information modification | 1792 | 2021-05-11 04:03:23 | | | | |
| 4 | Dean Liu | + 1 word(s) | 1793 | 2021-06-15 04:53:43 | | | | |
| 5 | Dean Liu | + 1 word(s) | 1793 | 2021-06-15 05:16:26 | | |
Video Upload Options
Hypereosinophilia (HE) is a heterogeneous condition with a persistent elevated eosinophil count of >350/mm3, which is reported in various (inflammatory, allergic, infectious, or neoplastic) diseases with distinct pathophysiological pathways. HE may be associated with tissue or organ damage and, in this case, the disorder is classified as hypereosinophilic syndrome (HES).
1. Eosinophil Development
Eosinophils are white blood cells of the granulocytic lineage that play an important role in innate immune functions [1] and develop in bone marrow from pluripotent stem cells expressing CD34+CD125+ antigens. These cells represent about 5% of the circulating blood leukocytes with an absolute eosinophil count (AEC) in healthy adults that is usually between 350 and 500/mm3, which increases during inflammatory processes, such as allergic diseases, parasitic, bacteria, and virus infection [2][3].
Structurally, they possess segmented bi-lobed nuclei and specific primary and secondary granules. Primary granules exhibit lysophospholipase activity that is involved in eosinophilic-dependent tissues inflammation [4]. Secondary granules contain many mediators, such as major basic protein (MBP), eosinophil cationic protein (ECP), eosinophil peroxidase (EPO), and eosinophil-derived neurotoxin (EDN), which are all able to induce both inflammation and tissue damage [5].
Furthermore, eosinophils are equipped of lipid bodies that play a critical role in asthma, as they cause eicosanoidis production [6]. Finally, they are potent productors of both reactive oxygen species and nitric oxide, which promote the anti-bacterial activity, while the ability to internalize the respiratory syncytial (RSV) and influenza viruses document the role of eosinophils in the viral response [7][8].
2. Eosinophil Contents, Biology and Homeostatic Immune Role
Detecting tissue-resident eosinophils showed that they are distributed in heart, skin, lung, and kidneys [9]. Despite this observation, under the homeostasis condition, eosinophils are particularly abundant in the gastrointestinal tract (GI), where they are involved in different biological processes. Both a beneficial and nonbeneficial role of eosinophils in GI tract have been postulated.
The first role is based on the ability of gastrointestinal eosinophils to mediate anti-parasitic response and promote with them a symbiotic association that is aimed at the maintenance of tissue homeostasis. Moreover, in the GI tract, a high number of eosinophils trapping bacteria represents an effective mechanism to protect this tissue from bacterial invasion.
The nonbeneficial role of gastrointestinal eosinophils is reported in Eosinophil Gastrointestinal Disorders (EGIDs) and Inflammation Bowel Disease (IBD). The pathogenesis of both diseases is dependent on tissue infiltration of eosinophils, followed by the accumulation of activated immune cells, such as B and T cells, as well as the production of pro-inflammatory cytokines [8].
An additional particular function of eosinophil is their role in tissue regeneration and remodeling. By the secretion of IL-4, eosinophils are able to facilitate liver and muscle regeneration [10][11], while the increased presence of these cells in the endometrium prompted speculation that they may play a role in tissue remodeling during ovulation and menstruation [12]. The absolute number and biology of eosinophils are both usually controlled by type-2 cytokines, such as interleukin-5 (IL-5), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3), produced by T-lymphocytes, mast cells, and stromal cells. IL-5, IL-3, and GM-CSF induce eosinophils maturation, survival, and apoptosis inhibition by PI3K, ERK, and STATs pathway activation. However, in this biological system, IL-5 shows a more prominent role than IL-3 and GM-CSF dependent on its high specificity for this leukocyte subset. In particular, the IL-5 receptor (IL-5R) plays a central role of intracellular signals regulation by its α and β-chains. The α-chain contains the ligand-binding subunit while the β-chain is defined as non-ligand-binding subunit and it mediates the intracellular signal transduction. The β-chain, in turn, is shared with IL-3 and GM-CSF receptors (IL-3R) (GM-CSFR), thus supporting the intriguing role of IL-5 as central mediator of type-2 cytokines-dependent eosinophil survival. An interestingly involvement of IL-5 concerns its production by group 2 innate lymphoid cells (ILC2s) after interleukin-33 (IL-33) stimulation. IL-33 promotes the IL-5 production from ILC2s, which, in turn, releasing IL-5 improves eosinophils expansion and survival. Hence, ILC2s are a new important mediator of IL-33-driven eosinophil disorder development [13][14].
Hence, IL-5, to date, is an attractive therapeutic target for the treatment of eosinophil-mediated disorders [15]. Nevertheless, despite these results, the published data report that IL-5 over-expression or down-regulation, alone, fail to induce eosinophil-mediated damage or eosinophils maturation [16], making its role unclear. However, all together, these observations implicate that the deregulation of IL-3, IL-5, and GM-CSF signaling may cause HES [15][17][18].
Finally, an important role in eosinophils biology concerns the fibrogenic activity of transforming growth factor beta-1 (TGF-B1). Eosinophils are a strong productor of TGF-B1, which is involved in airway remodeling or in disease state in different tissue, such as skin (atopy), nose (nasal polyposis), and blood (idiopathic hypereosinophilic syndrome) [19].
Eosinophils biology is also regulated by the cell surface glycan-binding protein, named siglecs (sialic acid immunoglobulin-like lectins). The most important glycan in eosinophils biology is Siglec-8 (originally named sialoadhesin family 2—SAF-2), as it is expressed selectively in these leukocyte types. Siglec-8 is stimulated by sialic acids, which induce the activation of two tyrosine-based motifs, defined immunoreceptor tyrosine-based inhibitor motif (ITIM) and a membrane-distal immunoreceptor tyrosine-based switch motif (ITSM), thus initiating the downstream receptor functions. Siglec-8 is involved in reactive oxygen species (ROS) production, in the loss of mitochondrial membrane potential, ERK1/2 activation, and caspase cleavage modulating apoptosis and cell survival [20]. For these regions, Siglec-8 was studied as a therapeutic target in patients with eosinophilic disorders using chimeric antibodies [21].
3. Eosinophil Recruitment into Blood and Tissue, Survival and Death
The term eosinophilia is employed for a small increase of the AEC in the blood (up to 1500/mm3), while hypereosinophilia (HE) indicates an AEC greater than 1500/mm3 on two consecutive blood samples drawn at a one-month interval. This persistent eosinophilia is usually linked to helminth infections, allergies, atopy, drugs, neoplastic disorders, or autoimmune diseases [22][23].
A second type of hypereosinophilia is tissue HE, which is defined as a percentage of eosinophils in the bone marrow (BM) that exceeds 20% of all nucleated cells, followed by extensive tissue infiltration, such as skin (in 69% of subjects), lung, and the gastrointestinal tract (44% and 38%, respectively) [22][23][24].
Upon activation, eosinophils infiltrate tissues, degranulate, and release proinflammatory cytokines that cause organ damage and dysfunction, defined as the Hypereosinophilic Syndrome (HES). HES represents a group of heterogeneous disorders characterized by persistent and unexplained HE in the blood or peripheral tissues usually associated with multiple organ damage or dysfunction. This damage may be due to direct cytotoxic effects of the eosinophil granulate contents or may occur because of the secondary involvement of other cell types [24][25]. Once eosinophils leave the blood circulation and migrate into tissue sites, they do not recirculate. Into the tissue, the survival is dependent on local production of cytokines that also prevent eosinophil apoptosis for several days. In fact, the patterns of cytokines regulate the recruitment of eosinophils to the specific tissue sites, activate endothelial cells, and induce tissue-resident cells to produce eosinophil-active chemokines to facilitate their preferential migration [26].
The cardiovascular system is often involved in HES [27][28].
The presenting symptoms of HE and HES are variable and they may include weakness, fatigue, cough, dyspnea and rhinitis, myalgias or angioedema, rash or fever, as well as severe tissue damage or end-organ failure [23][29]. Leukocytosis, anemia, abnormal platelet counts, increased vitamin B12 (>1000 pg/mL), and tryptase (>12 ng/mL) levels represent additional alterations that are associated with the disease [23][24].
Because HES is a rare neoplasm, its epidemiology has not been accurately investigated. Hence, the disease true incidence is unknown, which is mainly due to the lack of specific coding for the different HES variants. Although HES is mainly diagnosed in adults, 20 to 50 years old, it can also affect children and the elderly [30].
4. Classification of Hypereosinophilia and Hypereosinophilic Syndrome
HE and HES can usually be divided into multiple subgroups based upon clinical, laboratory, and molecular features.
HE is classified in three groups: primary (neoplastic or clonal), secondary (reactive), and idiopathic [24].
Th Working Conference on Eosinophil Disorder and Syndromes proposed various terminologies for eosinophilic syndromes. The HE subtypes were then divided into a hereditary (familial) variant (HEFA), HE of undetermined significance (HEUS), primary (clonal/neoplastic) HE produced by clonal/neoplastic eosinophils (HEN), and secondary (reactive) HE (HER), with the latter group including the lymphocyte variant as a subtype. The HEUS acronym was introduced as a novel term instead of idiopathic
HE [30]. The main two pathogenic forms of HES are the myeloproliferative (M-HES) and lymphocytic (L-HES) forms of the disease, respectively, classified as Myeloid Hypereosinophilic Syndrome and Lymphocytic Hypereosinophilic Syndrome. Each group includes several clinically distinct HES disorders [31]. Patients that do not display the M- or L-HES diagnosis can be classified as being affected by a Idiopathic Hypereosinophilic Syndrome or a Chronic Eosinophilic Leukemia non otherwise specified (CEL-NOS).
5. Molecular Pathogenesis in Hypereosinophilic Syndrome
The laboratory screening performed to formulate a HES diagnosis allow for us to understand molecular events that cause gene driver alterations in myeloid and lymphoid disorders that are associated with eosinophilia (summarized in Table 1).
Table 1. Molecular pathogenesis in Hypereosinophilic Syndrome
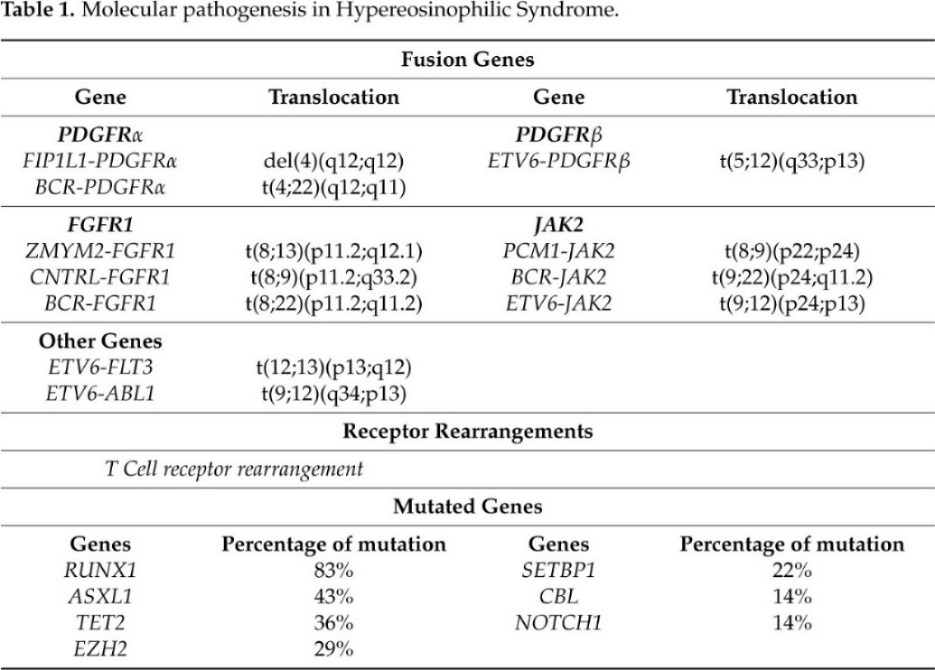
Nowadays, the T-cell receptor (TCR) gene rearrangements and Next Generation Sequencing (NGS) have been used to investigate the aberrant T-cell immunophenotype and somatic mutations in HES patients, respectively. Clonal TCR gene rearrangement is required for the diagnosis of most L-HES patients [32][33]. However, a study by Roufosse and colleagues demonstrated that not all HES patients exhibit a clonal TCR gene rearrangement than it is unclear whether such clonal T-cell population is always the cause of the disease [33].
Different studies based on NGS analysis reveled that the most frequently mutated genes are ASXL transcriptional regulator 1 (ASXL1) (43%), Tet methylcytosine dioxygenase 2 (TET2) (36%), Enhancer of zeste homolog 2(EZH2) (29%), SET binding protein 1 (SETBP1) (22%), Casitas B-lineage Lymphoma (CBL) (14%), and Notch homolog 1, translocation-associated (NOTCH1) (14%) [34] [35] [36].
6. Therapeutic Option for Patients with Eosinophilic Disorders
The best clinical treatment of HES depends on disease etiology and subtypes. However, even in the absence of a known cause, HES must be promptly treated in order to reduce potential morbidity that can result from organ damage. The main therapeutic options for HES patients can be divided in five groups: corticosteroids, cytotoxic agents, tyrosine kinase inhibitors, monoclonal antibodies (mAb), and chemotherapy (Figure 1).
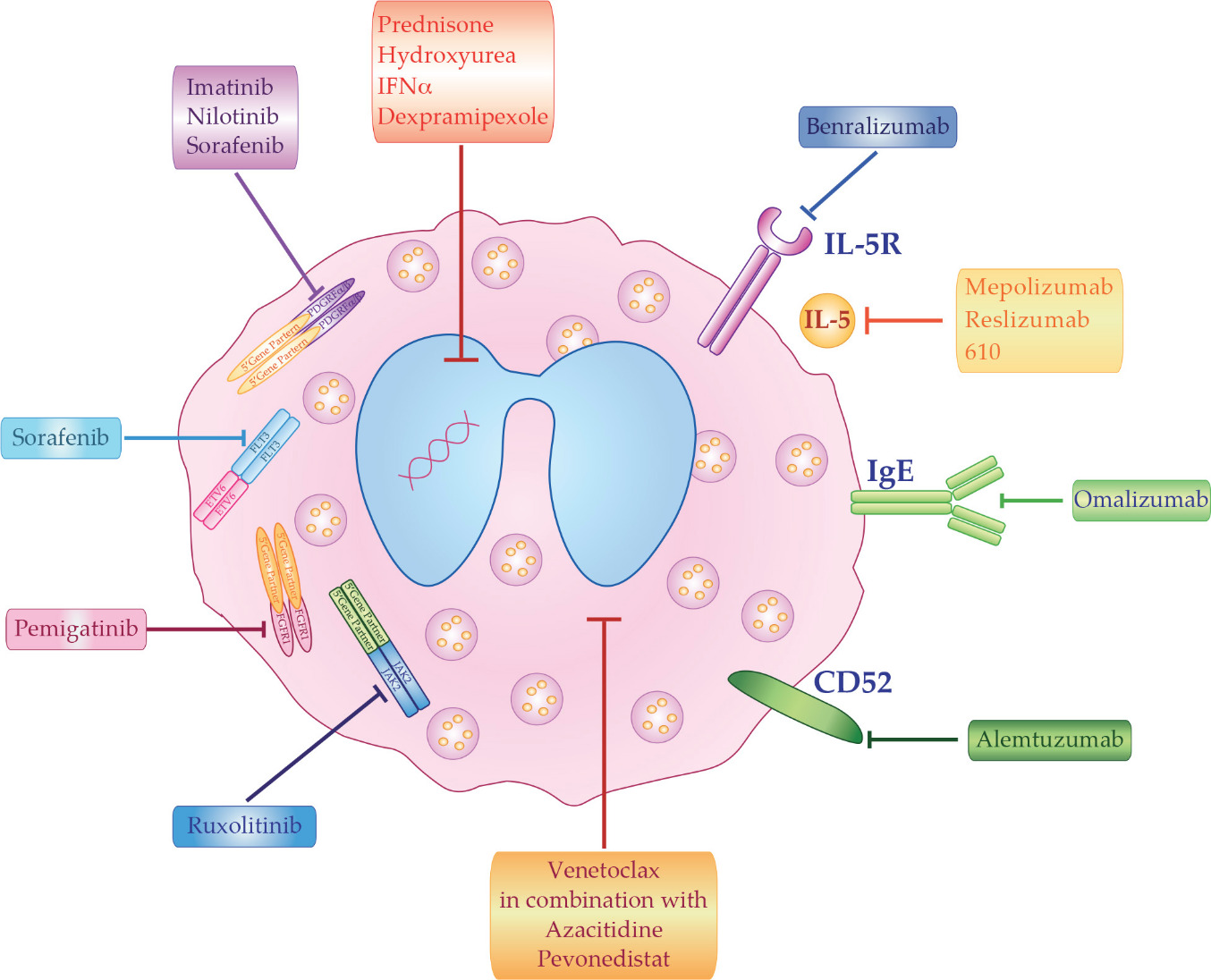
7. Conclusion
The distinction of each HES variant is critical for the appropriate management of the disease. The development of non-invasive sampling methods, coupled with an extensive NGS-based molecular characterization, will be important in distinguishing the different disease variants and discriminating an eosinophil myeloid neoplasia from HES. Moreover, this approach will enable an accurate disease monitoring, promptly identifying patients with a rapidly progressing hematological malignancy. Different ongoing clinical trials based on different drugs, used alone or in combination, will allow for a better understanding of the best initial therapy for any singlepatients with HES while taking the individual pathogenesis into consideration. Indeed,more targeted approach to treatment need an implementation of significative changes in the way that patients are managed through a more personalized approach to prognostication, the prediction of treatment responses.
References
- Wang, S.A.; Tam, W.; Tsai, A.G.; Arber, D.A.; Hasserjian, R.P.; Geyer, J.T.; George, T.I.; Czuchlewski, D.R.; Foucar, K.; Rogers, H.J.; et al. Targeted next-generation sequencing identifies a subset of idiopathic hypereosinophilic syndrome with features similar to chronic eosinophilic leukemia, not otherwise specified. Mod. Pathol. 2016, 29, 854–864.
- Lee, J.S.; Seo, H.; Im, K.; Park, S.N.; Kim, S.M.; Lee, E.K.; Kim, J.A.; Lee, J.H.; Kwon, S.; Kim, M.; et al. Idiopathic hypereosinophilia is clonal disorder? Clonality identified by targeted sequencing. PLoS ONE 2017, 12, e0185602.
- Simon, H.U.; Rothenberg, M.E.; Bochner, B.S.; Weller, P.F.; Wardlaw, A.J.; Wechsler, M.E.; Rosenwasser, L.J.; Roufosse, F.; Gleich, G.J.; Klion, A.D. Refining the definition of hypereosinophilic syndrome. J. Allergy Clin. Immunol. 2010, 126, 45–49.
- Maric, I.; Sun, X. Advances in diagnosis of mastocytosis and hypereosinophilic syndrome. Semin. Hematol. 2019, 56, 22–29.
- Helbig, G.; Wieczorkiewicz, A.; Dziaczkowska-Suszek, J.; Majewski, M.; Kyrcz-Krzemien, S. T-cell abnormalities are present at high frequencies in patients with hypereosinophilic syndrome. Haematologica 2009, 94, 1236–1241.
- Pardanani, A.; Lasho, T.; Wassie, E.; Finke, C.; Zblewski, D.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Predictors of survival in WHO-defined hypereosinophilic syndrome and idiopathic hypereosinophilia and the role of next-generation sequencing. Leukemia 2016, 30, 1924–1926.
- Wang, S.A.; Tam, W.; Tsai, A.G.; Arber, D.A.; Hasserjian, R.P.; Geyer, J.T.; George, T.I.; Czuchlewski, D.R.; Foucar, K.; Rogers, H.J.; et al. Targeted next-generation sequencing identifies a subset of idiopathic hypereosinophilic syndrome with features similar to chronic eosinophilic leukemia, not otherwise specified. Mod. Pathol. 2016, 29, 854–864.
- Lee, J.S.; Seo, H.; Im, K.; Park, S.N.; Kim, S.M.; Lee, E.K.; Kim, J.A.; Lee, J.H.; Kwon, S.; Kim, M.; et al. Idiopathic hypereosinophilia is clonal disorder? Clonality identified by targeted sequencing. PLoS ONE 2017, 12, e0185602.
- Weller, P.F.; Spencer, L.A. Functions of tissue-resident eosinophils. Nat. Rev. Immunol. 2017, 17, 746–760.
- Goh, Y.P.; Henderson, N.C.; Heredia, J.E.; Red Eagle, A.; Odegaard, J.I.; Lehwald, N.; Nguyen, K.D.; Sheppard, D.; Mukundan, L.; Locksley, R.M.; et al. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9914–9919.
- Heredia, J.E.; Mukundan, L.; Chen, F.M.; Mueller, A.A.; Deo, R.C.; Locksley, R.M.; Rando, T.A.; Chawla, A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 2013, 153, 376–388.
- Jeziorska, M.; Salamonsen, L.A.; Woolley, D.E. Mast cell and eosinophil distribution and activation in human endometrium throughout the menstrual cycle. Biol. Reprod. 1995, 53, 312–320.
- Choi, H.S.; Won, T.; Hou, X.; Chen, G.; Bracamonte-Baran, W.; Talor, M.V.; Jurcova, I.; Szarszoi, O.; Curnova, L.; Striz, I.; et al. Innate Lymphoid Cells Play a Pathogenic Role in Pericarditis. Cell Rep. 2020, 30, 2989–3003.e6.
- Johansson, K.; Malmhall, C.; Ramos-Ramirez, P.; Radinger, M. Bone marrow type 2 innate lymphoid cells: A local source of interleukin-5 in interleukin-33-driven eosinophilia. Immunology 2018, 153, 268–278.
- Roufosse, F. Targeting the Interleukin-5 Pathway for Treatment of Eosinophilic Conditions Other than Asthma. Front. Med. 2018, 5, 49.
- Dent, L.A.; Strath, M.; Mellor, A.L.; Sanderson, C.J. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 1990, 172, 1425–1431.
- Esnault, S.; Kelly, E.A. Essential Mechanisms of Differential Activation of Eosinophils by IL-3 Compared to GM-CSF and IL-5. Crit. Rev. Immunol. 2016, 36, 429–444.
- Dougan, M.; Dranoff, G.; Dougan, S.K. GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 2019, 50, 796–811.
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4, 93.
- O’Sullivan, J.A.; Carroll, D.J.; Bochner, B.S. Glycobiology of Eosinophilic Inflammation: Contributions of Siglecs, Glycans, and Other Glycan-Binding Proteins. Front. Med. 2017, 4, 116.
- Legrand, F.; Cao, Y.; Wechsler, J.B.; Zhu, X.; Zimmermann, N.; Rampertaap, S.; Monsale, J.; Romito, K.; Youngblood, B.A.; Brock, E.C.; et al. Sialic acid-binding immunoglobulin-like lectin (Siglec) 8 in patients with eosinophilic disorders: Receptor expression and targeting using chimeric antibodies. J. Allergy Clin. Immunol. 2019, 143, 2227–2237.
- Valent, P.; Klion, A.D.; Horny, H.P.; Roufosse, F.; Gotlib, J.; Weller, P.F.; Hellmann, A.; Metzgeroth, G.; Leiferman, K.M.; Arock, M.; et al. Contemporary consensus proposal on criteria and classification of eosinophilic disorders and related syndromes. J. Allergy Clin. Immunol. 2012, 130, 607–612.
- Shomali, W.; Gotlib, J. World Health Organization-defined eosinophilic disorders: 2019 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2019, 94, 1149–1167.
- Mattis, D.M.; Wang, S.A.; Lu, C.M. Contemporary Classification and Diagnostic Evaluation of Hypereosinophilia. Am. J. Clin. Pathol. 2020.
- Schwaab, J.; Jawhar, M.; Naumann, N.; Schmitt-Graeff, A.; Fabarius, A.; Horny, H.P.; Cross, N.C.; Hofmann, W.K.; Reiter, A.; Metzgeroth, G. Diagnostic challenges in the work up of hypereosinophilia: Pitfalls in bone marrow core biopsy interpretation. Ann. Hematol. 2016, 95, 557–562.
- Ackerman, S.J.; Bochner, B.S. Mechanisms of eosinophilia in the pathogenesis of hypereosinophilic disorders. Immunol. Allergy Clin. N. Am. 2007, 27, 357–375.
- Lefevre, G.; Copin, M.C.; Staumont-Salle, D.; Avenel-Audran, M.; Aubert, H.; Taieb, A.; Salles, G.; Maisonneuve, H.; Ghomari, K.; Ackerman, F.; et al. The lymphoid variant of hypereosinophilic syndrome: Study of 21 patients with CD3-CD4+ aberrant T-cell phenotype. Medicine 2014, 93, 255–266.
- Leru, P.M. Eosinophilic disorders: Evaluation of current classification and diagnostic criteria, proposal of a practical diagnostic algorithm. Clin. Transl. Allergy 2019, 9, 36.
- Weller, P.F.; Bubley, G.J. The idiopathic hypereosinophilic syndrome. Blood 1994, 83, 2759–2779.
- Swerdlow, S.H.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. In WHO Classification of Tumours, Revised, 4th ed.; IARC Pubblication: Lyon, France, 2017; Volume 2.




