
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hsiuying Wang | + 3360 word(s) | 3360 | 2021-04-25 07:52:49 | | | |
| 2 | Peter Tang | Meta information modification | 3360 | 2021-04-27 05:18:04 | | |
Video Upload Options
Parkinson’s disease (PD) is a neurodegenerative disorder that affects 1% of the population over the age of 60. Diabetes Mellitus (DM) is a metabolic disorder that affects approximately 25% of adults over the age of 60. Recent studies showed that DM increases the risk of developing PD. The link between DM and PD has been discussed in the literature in relation to different mechanisms including mitochondrial dysfunction, oxidative stress, and protein aggregation.
1. Introduction
Parkinson’s disease (PD) is a chronic neurodegenerative disease that has an increasing prevalence with age [1]. PD affects 1% of the population above 60 years and is called early-onset PD if it begins before age 50. The non-motor symptoms of PD include sleep disorders, depression, cognitive changes, illusions, and delusions [2]. The motor symptoms of PD include tremor, slowed movement, rigid muscles, impaired posture and balance, speech changes, and writing changes [3]. Increasing evidence shows that biological sex is an important factor in the development of PD. The relationship between estrogen exposure and PD risk was investigated, and women with higher cumulative estrogen exposure had a significantly reduced PD risk [4][5].
Several factors can modify the risk of developing PD. The increasing risk factors include pesticides, consumption of dairy products, history of melanoma, and traumatic brain injury, whereas the decreasing risk factors include smoking, caffeine consumption, higher serum urate concentrations, physical activity, and use of ibuprofen and other common medications [6].
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is a drug that can cause irreversible parkinsonism [7]. In addition, metabolic syndrome may be a risk factor for PD development [8][9]. The stimulation of oxidative stress is pivotal to the evolution of metabolic syndrome and PD [10][11]. Besides, a recent study showed the potential link between gut microbiota and PD [12]. It has been proposed that the neurodegenerative cascade may be initiated in the gut with subsequent spreading to the brain and that gut microbiota could be involved in this process.
In recent years, an emerging body of evidence has shown the association between PD and diabetes mellitus (DM). The cause of DM is a result of either the pancreas not producing enough insulin or the body not responding appropriately to insulin. Hyperglycemia affects people who have DM, and chronic hyperglycemia is associated with long-term damage and dysfunction of different organs [13]. According to the diabetes website (https://www.who.int/health-topics/diabetes#tab=tab_1) of the World Health Organization on 1 March 2021, about 422 million people worldwide have DM in 2021. Both the number of cases and the prevalence of DM have been increasing over the past few decades.
There are two main types of DM: type 1 and type 2. In type 1 DM, the pancreas fails to produce enough insulin. People with type 1 DM must use insulin injections to control their blood glucose. In type 2 DM, the pancreas produces insulin, but the body does not respond appropriately to insulin.
Most people with type 2 DM are obese, and obesity may cause insulin resistance [13]. Patients with DM may have many complications, including retinopathy, nephropathy, peripheral neuropathy, and autonomic neuropathy. There is also an increased incidence of hypertension, atherosclerotic cardiovascular complications, peripheral arterial complications, and abnormalities of lipoprotein metabolism. Diabetic retinopathy (DR) is a common complication of DM, and diabetic kidney disease (DKD) or diabetic nephropathy is a type of chronic kidney disease caused by DM. DKD was reported in approximately 40% of DM patients, and the majority of DKD patients die from cardiovascular diseases and infections [14].
The diagnosis of DM is based on fasting sugar blood tests or A1c blood tests. Compared with the simplicity of DM diagnosis, it is more difficult to conclusively diagnose PD due to the lack of a reference standard test [15]. The diagnosis of PD is based on a review of patients’ signs and symptoms, and neurological and physical examinations. The genetic factor can be identified in 5–10% of the patients. Studies show that PD is associated with five genes: α-synuclein (SNCA); parkin (PARK2); PTEN-induced putative kinase 1 (PINK1); DJ-1 (PARK7); and Leucine-rich repeat kinase 2 (LRRK2) [16][17][18][19]. In addition, a meta-analysis on genome-wide association studies (GWAS) from 13,708 cases and 95,282 controls has identified 28 independent risk alleles at 24 gene loci associated with a risk for PD [20]. The gene expression differences between PD and healthy controls can be used as a potential prognosis of PD. In addition to the gene biomarker, the circulating microRNA (miRNA) can be a useful biomarker for PD as well as DM.
2. MicroRNA
miRNA is a small, non-coding RNA about 21–24 nucleotides in length that has important functions in cell differentiation, development, the regulation of the cell cycle, and apoptosis. The first miRNA was discovered in the early 1990s when studying the nematode Caenorhabditis elegans regarding the gene lin-14 [21]. miRNAs play an important epigenetic role involved in many diseases and can be overexpressed or repressed in different diseases. The inhibition or replacement of miRNAs is a promising area of study for therapeutics [22].
The biogenesis of miRNA is classified into canonical and non-canonical pathways. Most miRNAs are transcribed from DNA sequences into primary miRNAs (pri-miRNAs) and processed into precursor miRNAs (pre-miRNAs) and mature miRNAs. miRNAs are synthesized from primary miRNAs in two stages by the action of two RNase III-type proteins [23][24]. miRNAs may regulate up to 30% of the protein-coding genes in the human genome [25] and are well known to be involved in the initiation and progression of cancers [26][27][28][29][30][31][32][33]. In addition to being tumor suppressors or oncogenes of cancer, miRNAs also contribute to neurological diseases. Let-7b is a miRNA biomarker for anti-NMDA receptor encephalitis [34][35][36]. For neurological diseases, miRNAs were identified to account for PD, amyotrophic lateral sclerosis, frontotemporal dementia, Alzheimer’s disease, spinal muscular atrophy, Prader–Willi syndrome, Niemann–Pick disease, neurofibromatosis, narcolepsy, Friedreich’s ataxia, and ataxia-telangiectasia [36][37][38][39][40][41]. miRNAs are also explored as being related to DM [42].
3. MicroRNA Biomarkers
I collected some common miRNA biomarkers of PD and DM from the literature to discuss the association between both diseases. The common biomarkers are listed in Table 1.
Table 1. Common microRNAs (miRNAs) related to Parkinson’s disease (PD) and diabetes mellitus (DM).
|
microRNA |
Parkinson’s Disease Reference |
Diabetes Reference |
|---|---|---|
|
miR-92a |
||
|
miR-100 |
||
|
miR-23a |
||
|
let-7 |
[38] |
[53] |
|
miR-485 |
[55] |
|
|
miR-26 |
||
|
miR-146a |
||
|
miR-335-3p |
[60] |
|
|
miR-155 |
||
|
miR-1 |
||
|
miR-19b-3p |
||
|
miR-153 |
[70] |
|
|
miR-409-3p |
[70] |
|
|
miR-10a-5p |
[70] |
[81] |
|
let-7g-3p |
[70] |
|
|
miR-103a-3p |
[65] |
|
|
miR-200 |
[65] |
|
|
miR-204 |
||
|
miR-21 |
||
|
miR-96 |
||
|
miR-17 |
[105] |
|
|
miR-365 |
[56] |
|
|
miR-18a |
[44] |
|
|
miR-125a |
[112] |
|
|
miR-125b |
[115] |
|
|
miR-10b |
||
|
miR-200c |
||
|
miR-210 |
[125] |
|
|
miR-218 |
||
|
miR-195 |
[54] |
|
|
miR-7 |
||
|
miR-148a |
[54] |
|
|
miR-182 |
[137] |
|
|
miR-34a |
||
|
miR-133b |
[143] |
|
|
miR-145 |
||
|
miR-143 |
[56] |
|
|
miR-342 |
[149] |
|
|
miR-26b |
||
|
miR-135b |
[154] |
|
|
miR-22 |
||
|
miR-20a |
||
|
miR-766 |
[160] |
[122] |
|
miR-30b |
[98] |
|
|
miR-30c |
||
|
miR-148b |
||
|
miR-29a |
||
|
miR-29c |
[165] |
[167] |
|
miR-1249 |
||
|
miR-18b |
||
|
miR-15a |
[56] |
|
|
miR-30a |
||
|
miR-9 |
[176] |
|
|
miR-132 |
[56] |
|
|
miR-423 |
||
|
miR-486 |
[56] |
|
|
miR-1260 |
[56] |
These two diseases share more miRNA biomarkers than those listed in Table 1. In this paper, we review miRNAs listed in Table 1 to connect PD and DM. The miR-92a, miR-100, and miR-23a were shown to significantly target 244 gene biomarkers of PD that were identified by integrating three datasets including 35 normal control and 25 PD patients’ substantia nigra mRNA expression profiles [43]. The tripartite regulatory network identified miR-18a, -92a, -200a, -200c, -17, and -20a as hub miRNAs that can be considered as possible biomarkers for PD [44]. miR-92a may serve to correct diabetes-associated inflammation and restore normal circadian function in CD34+ cells [45]. miR-100-5p, miR-146a-5p, miR-148a-3p, miR-210-5p, and miR-342-3p were dysregulated in type 1 DM patients compared to controls [65]. In comparison with either normal glucose tolerance or type 2 DM subjects, miR-18a, miR-18b, and miR-23a decreased in impaired glucose tolerance subjects [51]. miR-30, miR-29, let-7, miR-485, and miR-26 were shown to be implicated in PD pathogenesis [38]. Let-7 could be involved in regulating neuronal degeneration in PD and let-7g-3p was up-regulated in the CSF of PD patients [38]. Let-7 regulated multiple aspects of glucose metabolism, and anti-miR-induced let-7 knockdown was suggested as a potential treatment for type 2 DM [53]. Microarray analysis of PD substantia nigra samples revealed that miR-485-5p and miR-204-5p were up-regulated and miR-155-5p and miR-423 were down-regulated. In addition, miR-200, miR-21, miR-195, miR-7, miR-148a, miR-145, miR-26b, and miR-135b also have different expression in PD samples compared to control samples [54]. Overexpression of miR-485 suppressed high glucose-induced proliferation of human mesangial cells [55]. miR-26a in β cells alleviated obesity-induced insulin resistance and hyperinsulinemia, and prevented hyperinsulinemia through targeting several critical regulators of insulin secretion and β cell proliferation [58]. A systematic review of literature summarized miRNAs as differing significantly between individuals with PD and healthy controls and/or between treated and untreated patients with PD including down-regulated miRNAs, miR-30b, miR-30c, miR-26a, miR-148b, miR-1, miR-22, miR-29a, miR-103a-3p, miR-1249, miR-20a, miR-18b, miR-15a, miR-143, miR-19b, and up-regulated miRNAs, miR-30a, miR-7, miR-9, miR-132, miR-423, miR-365, miR-486, miR-1260, and miR-218 [56]. miR-26a could ameliorate bone-specific insulin resistance and bone quality in diabetic mice, which depend on the insulin receptors on osteoblasts [57].
miR-146a, miR-335-3p, and miR-335-5p were down-regulated in idiopathic PD patients and patients with a mutation in the LRRK2 gene versus controls [60]. miR-146a-5p was down-regulated in recently diagnosed type 1 DM patients [65]. miR-21-5p, miR-100-5p, miR-148a, miR-146a-5p, miR-210-5p, and miR-342-3p were dysregulated in type 1 DM patients compared to controls [48]. The expression of miR-335 was negatively correlated with the secretion index in human islets of individuals with prediabetes [66]. An animal study explored the potential involvement of miR-155 in the pathogenesis of diabetes complications [68]. Except for the liver, the miR-155 expression level was significantly decreased in the diabetic kidney, heart, aorta, peripheral blood mononuclear cells, and the sciatic nerve versus the controls. miR-1 and miR-19b-3p showed decreased expression in PD, whereas miR-153, miR-409-3p, miR-10a-5p, and let-7g-3p were found to be up-regulated [70]. Type 2 DM patients expressed decreased levels of miR-1-3p and miR-34a-5p compared with controls [73]. miR-19b targets PD-related genes [74]. The long non-coding RNA maternally expressed gene 3 (MEG3) inhibited high glucose-induced apoptosis and inflammation by regulating the miR-19b/SOCS6 axis through the JAK2/STAT3 signaling pathway in the human retinal microvascular endothelial cells [185]. The miR-153 expression level was increased in IL-1β-treated β cells and primary islets from the diabetic rodents [76]. An insulin resistance group presented a remarkably higher serum miR-409-5p level than a non-insulin resistance group [79]. Acarbose, an α-glucosidase inhibitor, can regulate glucose metabolism through the MAPK pathway and can suppress proinflammatory cytokines by increasing miR-10a-5p and miR-664 in the ileum. Acarbose reduced blood glucose by activating miR-10a-5p in diabetic rats [81]. Let-7g was differentially expressed in patients with or at risk of for type 1 DM [82]. Significant overexpression of miR-103a-3p, miR-30b-5p, and miR-29a-3p was observed in treated patients with PD [84]. miR-21-5p, miR-103a-3p, miR-148b-3p, miR-155-5p, miR-200a-3p, and miR-210-3p were up-regulated in recently diagnosed type 1 DM patients compared with controls [65]. Serum miR-204 was elevated in children and adults with type DM [87].
miR-96-5p was involved in oxidative stress in PD [100]. The overexpression of miR-96 was found to lead to an impairment of insulin signaling and glycogen synthesis in hepatocytes [101]. A significant decrease in miR-17-3p in diabetic retinopathy as well as in proliferative diabetic retinopathy patients was shown when compared with non-diabetic retinopathy patients [105]. miR-125b-5p and miR-365a-3p have strong positive correlations with HbA1c [106]. A study proposed miRNA biomarker panels that efficiently distinguish early-stage PD patients from controls and miR-125a-5p and miR-10b-5p were identified in these miRNA panels [112]. miR-125a is overexpressed in insulin target tissues in a spontaneous rat model of type 2 DM [114]. miR-125b-5p is a putative target gene of the long non-coding RNA brain-derived neurotrophic factor anti-sense that might act as a potential therapeutic target for PD [115]. A titer of islet autoantibodies IAA was negatively associated with miR-10b-5p [122]. miR-200c-3p was down-regulated in the echinomycin-treated PD cellular model [123]. miR-200c-3p was positively correlated with HbA1c [106]. miR-210 target genes were identified to have a significant age-related neurodegenerative disease pathway enrichment including Huntington’s disease, Alzheimer’s disease, and PD [125]. Plasma miR-210 was significantly up-regulated in type 2 DM subjects in contrast to controls [126]. The down-regulation of miR-218 in the brain was related to PD via activation of NF-κB signaling [127]. Glucose up-regulated miR-218 expression, and miR-218 and RUNX2 might be vital targets for use in diagnosing and treating DR [129]. miR-195 was up-regulated in the frontal cortex region of the PD brain [54]. miR-195-5p expression was significantly increased in serum samples from gestational DM patients as compared with those in healthy pregnancies [133]. Serum miR-7 was significantly elevated in the type 2 DM patients and the type 2 DM-associated microvascular complications patients when compared with the controls [135]. Levels of miR-148a-3p were associated with glycemic status and glucose levels [111]. miR-182-5p mediates nigrostriatal protection in the MPTP model of PD [137]. miR-182-5p was very highly expressed in individuals with prediabetes or type 2 DM, and miR-182-5p was observed to be significantly under-expressed in type 2 DM relative to prediabetes [140].
miR-34a was differentially expressed in 1-methyl-4-phenylpyridinium (MPP)+-treated differentiated PC12 cells as a model of PD [142]. miR-34a and miR-9 were up-regulated in MPP+-treated differentiated PC12 cells as a model of PD [142]. miR-34a was increased in type 2 DM patients who were overweight and obese, and miR-34a was differentially affected by glycemia, obesity, insulin treatment, and the presence of nephropathy and diabetic foot [63]. The plasma level of miR-133b was reduced in PD patients compared with the controls [143]. Myocardial-specific miR-133b was confirmed to be down-regulated in diabetic rat hearts [71]. By comparing the miRNAs identified in this experiment with those previously reported to be associated with DKD, miR-133b was up-regulated in urinary exosomes in patients with type 2 DKD [144]. miR-145-3p in the PD group was higher than that in the control group [145]. In rats with type 1 DM, the therapeutic effects of stroke treatment were compared between bone-marrow stromal cells (BMSCs) derived from type 1 DM rats (DM-BMSCs) and BMSCs derived from normal rats (Nor-BMSCs). In vivo, compared with Nor-BMSC or phosphate-buffered saline treatment, DM-BMSC treatment improved functional outcome, decreased serum miR-145 expression, and increased expression of the miR-145 target genes ABCA1 and IGFR1 [146]. miR-342-3p, miR-29a-5p, and miR-9-5p were identified to regulate genes associated with PD such as CTSB and SPPL2B [149]. The level of miR-26b targeting hsc70 was significantly increased in PD substantia nigra pars compacta relative to actin mRNA levels [150]. miR-26b-5p was found to be significantly down-regulated following metformin treatments in patients with type 2 DM [97]. Ectosomes (Ects) are a subpopulation of extracellular vesicles, and the level of miR-26b-5p was significantly different between Ects obtained from patients with type 2 DM and those obtained from healthy controls [151]. miR-26b was detected in the blood of type 2 DM samples that indicated miR-26b as a promising biomarker of type 2 DM [152].
Both in vitro and in vivo, the expression of miR-135b decreased in retinal cells under hyperglycemia exposure and increased in the DM retina [154].
The dysregulations of miR-22 and miR-23a were shown in the comparison between PD patients and control individuals [50]. Three experimental rat groups were analyzed in a study: rats receiving a standard diet (N), rats receiving a high-fat diet (HFD), and rats receiving a high-fat diet (HFD) with simultaneous administration of T2 (HFD-T2). An approximate 50% decrease in the level of miR-22-3p was detected in the serum of HFD-T2 rats in comparison to HFD rats [155]. miR-20a-5p was significantly decreased in women with gestational DM compared with controls [159]. miR-766 could target the gene GBA, and mutations of GBA were the most common genetic risk factor for PD [160]. The blood glucose concentration measured at 120 min of an oral glucose tolerance test was correlated negatively to miR-766-3p [122]. miR-30b-5p was differentially expressed in PD [70]. mRNA and protein profiling of extracellular vesicles extracted from diabetic subjects with the DR group or without the DR group and healthy controls were performed. Modulation of miR-30b-5p inside microvascular cells confirmed their involvement in abnormal angiogenesis [98]. Serum miR-30c-5p levels correlate with disease duration in both multiple system atrophy and PD [161].
miR-30c and miR-148b were down-regulated in PD [162]. miR-30c reduced plasma cholesterol in several diet-induced and diabetic hypercholesterolemic mice [163]. The levels of HbA1c were negatively associated with miR-30c-5p [122]. miR-148b was detected in the blood of type 2 DM samples indicating miR-148b as a promising biomarker of type 2 DM [152]. The level of miR-148b was detected in the sera of healthy controls, individuals with impaired glucose regulation, and type 2 DM patients by real-time polymerase chain reaction (PCR). Compared with those in the healthy control group, the miR-148b level in the impaired glucose regulation group was significantly higher. miR-148b was also significantly higher in the type 2 DM group compared with the other groups [164]. The expression of serum miR-29a and miR-29c expression tended to decrease with PD severity [165]. In the liver, both miR-29a and miR-29c were important negative regulators of insulin signaling via phosphatidylinositol 3-kinase regulation [166]. Evidence showed that miR-29a and miR-29c were increased in skeletal muscle from patients with type 2 DM [167]. miR-1249 was altered in PD with a focus on early-onset PD and late-onset PD patients [168]. A set of miRNAs including miR-18b and miR-1249 inverted their trend after deep brain stimulation treatment, becoming down-regulated compared to PD untreated patients’ samples [170]. miR-1249 was differentially expressed in pre-DM, obese, and non-diabetic individuals at follow-up [82]. miR-1249 was associated with the DM complication nephropathy [169]. The long non-coding RNA KCNQ1OT1 that regulated miR-18b-5p could affect cell proliferation, apoptosis, and fibrosis in diabetic nephropathy [171]. The peripheral blood miR-15a expression levels were significantly decreased in patients with type 2 DM and pre-diabetes individuals exhibiting impaired fasting glucose and impaired glucose tolerance individuals, compared with healthy control subjects [172]. Expression of miR-15a was increased in skeletal muscle obtained from the gestational DM group and type 1 DM group compared with a control group of offspring from the background population [173].
An up-regulation trend was observed for miR-30a-5p in L-dopa-treated PD patients [175]. miR-30a-5p and miR-30c-5p were found to be involved in blood coagulation, platelet activation, glucose metabolism, insulin signaling, and inflammation and were significantly up-regulated in type 2 DM [126]. miR-30a-5p is associated with dysglycaemia and could potentially predict prediabetes [140]. Deregulated plasma levels of miR-30a-5p were observed years before the onset of type 2 DM and pre-DM and could be used to evaluate the risk of developing DM [174]. miR-9-5p was one of several miRNAs that might target 13 genes associated with PD [149]. Experiments based on islet cell lines indicated that the overexpression of miR-9 decreased glucose-stimulated insulin secretion, while the knockdown of miR-9 promotes insulin secretion to a certain degree [176]. Expression of miR-132 was decreased in serum and placenta tissues in gestational DM patients compared with the healthy women [177]. miR-132 played a critical role in the regeneration of mouse islet β cells through the down-regulation of its target Pten, and the miR-132/Pten/Akt/Foxo3 signaling pathway might represent a suitable target to enhance β cell mass [178]. Lowered miR-423 levels in DM patients showed a correlation with vascular endothelial growth factor and an inverse correlation between nitric oxide and endothelial nitric oxide synthase expression. Hence, miR-423 may be involved in the regulation of diabetic vascular retinal proliferation [179]. miR-486-5p identified responders to thiazolidinedione therapy among the insulin-resistant group [181]. The serum levels of miR-486 were significantly reduced in patients with DKD when compared with the healthy control and type 2 DM groups [182]. In the comparison of type 1 DM versus type 2 DM, miR-1260 was differently expressed in the two groups [183]. Circulating miR-1260a was differently expressed at two time points in elderly type 2 DM patients who did not respond to sitagliptin treatment [184].
In summary, the miRNAs in Table 1 were involved in the functions that are related PD or DM. Figure 1 shows some miRNA-related functions that may cause PD or DM.
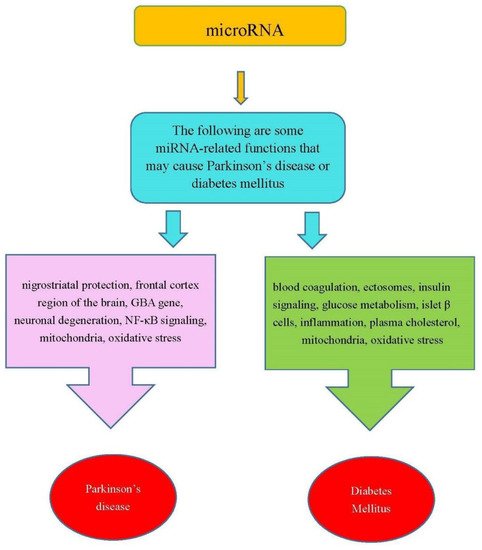
Figure 1. Some miRNA-related functions may cause PD and DM.
References
- Tysnes, O.B.; Storstein, A. Epidemiology of Parkinson’s disease. J. Neural Transm. 2017, 124, 901–905.
- Fénelon, G.; Mahieux, F.; Huon, R.; Ziégler, M. Hallucinations in Parkinson’s disease: Prevalence, phenomenology and risk factors. Brain 2000, 123, 733–745.
- Poluha, P.C.; Teulings, H.L.; Brookshire, R.H. Handwriting and speech changes across the levodopa cycle in Parkinson’s disease. Acta Psychol. 1998, 100, 71–84.
- Haaxma, C.A.; Bloem, B.R.; Borm, G.F.; Oyen, W.J.; Leenders, K.L.; Eshuis, S.; Booij, J.; Dluzen, D.E.; Horstink, M.W. Gender differences in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2007, 78, 819–824.
- Cerri, S.; Mus, L.; Blandini, F. Parkinson’s disease in women and men: What’s the difference? J. Parkinson’s Dis. 2019, 9, 501–515.
- Ascherio, A.; Schwarzschild, M.A. The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol. 2016, 15, 1257–1272.
- Langston, J.W. The MPTP Story. J. Parkinsons Dis. 2017, 7, S11–S19.
- Zhang, P.; Tian, B. Metabolic syndrome: An important risk factor for Parkinson’s disease. Oxid. Med. Cell Longev. 2014, 2014, 729194.
- Nam, G.E.; Kim, S.M.; Han, K.; Kim, N.H.; Chung, H.S.; Kim, J.W.; Han, B.; Cho, S.J.; Yu, J.H.; Park, Y.G.; et al. Metabolic syndrome and risk of Parkinson disease: A nationwide cohort study. PLoS Med. 2018, 15, e1002640.
- Whaley-Connell, A.; McCullough, P.A.; Sowers, J.R. The role of oxidative stress in the metabolic syndrome. Rev. Cardiovasc. Med. 2011, 12, 21–29.
- Zhou, C.; Huang, Y.; Przedborski, S. Oxidative stress in Parkinson’s disease: A mechanism of pathogenic and therapeutic significance. Ann. N. Y. Acad. Sci. 2008, 1147, 93–104.
- Weis, S.; Schwiertz, A.; Unger, M.M.; Becker, A.; Faßbender, K.; Ratering, S.; Kohl, M.; Schnell, S.; Schäfer, K.H.; Egert, M. Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Parkinsons Dis. 2019, 5, 28.
- American Diabetes, A. Diagnosis and classification of diabetes mellitus. Diabetes Care 2013, 36 (Suppl. 1), S67–S74.
- Alicic, R.Z.; Rooney, M.T.; Tuttle, K.R. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin. J. Am. Soc. Nephrol. 2017, 12, 2032–2045.
- Levine, C.B.; Fahrbach, K.R.; Siderowf, A.D.; Estok, R.P.; Ludensky, V.M.; Ross, S.D. Diagnosis and treatment of Parkinson’s disease: A systematic review of the literature. Evid. Rep. Technol. Assess Summ. 2003, 57, 1–4.
- Zimprich, A.; Biskup, S.; Leitner, P.; Lichtner, P.; Farrer, M.; Lincoln, S.; Kachergus, J.; Hulihan, M.; Uitti, R.J.; Calne, D.B.; et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004, 44, 601–607.
- Gasser, T. Usefulness of Genetic Testing in PD and PD Trials: A Balanced Review. J. Parkinsons Dis. 2015, 5, 209–215.
- Stefanis, L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect Med. 2012, 2, a009399.
- Dawson, T.M.; Dawson, V.L. The role of parkin in familial and sporadic Parkinson’s disease. Mov. Disord 2010, 25 (Suppl. 1), S32–S39.
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993.
- Lee, R.; Feinbaum, R.; Ambros, V. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139.
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20.
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004.
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610.
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Dutkiewicz, A.; Giefing, M.; Lewandowska, M.A.; Kozlowski, P. Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants. Cancers 2019, 11, 793.
- Xian, Q.J.; Zhao, R.L.; Fu, J.J. MicroRNA-527 Induces Proliferation and Cell Cycle in Esophageal Squamous Cell Carcinoma Cells by Repressing PH Domain Leucine-Rich-Repeats Protein Phosphatase 2. Dose Response 2020, 18.
- Chen, X.; Zhang, Z.; Ma, Y.; Su, H.; Xie, P.; Ran, J. LINC02381 Promoted Cell Viability and Migration via Targeting miR-133b in Cervical Cancer Cells. Cancer Manag. Res. 2020, 12, 3971–3979.
- Wang, H. Phylogenetic Analysis to Explore the Association Between Anti-NMDA Receptor Encephalitis and Tumors Based on microRNA Biomarkers. Biomolecules 2019, 9, 572.
- Wang, H. Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis. Int. J. Mol. Sci. 2016, 17, 773.
- Wang, H. Predicting Cancer-Related MiRNAs Using Expression Profiles in Tumor Tissue. Curr. Pharm. Biotechnol. 2014, 15, 438–444.
- Zhang, J.; Xu, X.; Zhao, S.; Gong, Z.; Liu, P.; Guan, W.; He, X.; Wang, T.; Peng, T.; Teng, J. The Expression and Significance of the Plasma Let-7 Family in Anti-N-methyl-D-aspartate Receptor Encephalitis. J. Mol. Neurosci. 2015, 56, 531–539.
- Wang, H. Efficacies of treatments for anti-NMDA receptor encephalitis. Front. Biosci. 2016, 21, 651–663.
- Taguchi, Y.H.; Wang, H. Exploring microRNA Biomarker for Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2018, 19, 1318.
- Rizzuti, M.; Filosa, G.; Melzi, V.; Calandriello, L.; Dioni, L.; Bollati, V.; Bresolin, N.; Comi, G.P.; Barabino, S.; Nizzardo, M.; et al. MicroRNA expression analysis identifies a subset of downregulated miRNAs in ALS motor neuron progenitors. Sci. Rep. 2018, 8, 1–12.
- Goh, S.Y.; Chao, Y.X.; Dheen, S.T.; Tan, E.K.; Tay, S.S. Role of MicroRNAs in Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 5649.
- Grasso, M.; Piscopo, P.; Talarico, G.; Ricci, L.; Crestini, A.; Tosto, G.; Gasparini, M.; Bruno, G.; Denti, M.A.; Confaloni, A. Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiol. Aging 2019, 84, 240 e1–240 e12.
- Magri, F.; Vanoli, F.; Corti, S. mi RNA in spinal muscular atrophy pathogenesis and therapy. J. Cell. Mol. Med. 2018, 22, 755–767.
- Wang, H. Phylogenetic Analysis of microRNA Biomarkers for Amyotrophic Lateral Sclerosis. Biocell 2021, 45, 547–561.
- Wang, H. MicroRNA, Diabetes Mellitus and Colorectal Cancer. Biomedicines 2020, 8, 530.
- Taguchi, Y.; Wang, H. Exploring MicroRNA Biomarkers for Parkinson’s Disease from mRNA Expression Profiles. Cells 2018, 7, 245.
- Chatterjee, P.; Bhattacharyya, M.; Bandyopadhyay, S.; Roy, D. Studying the system-level involvement of microRNAs in Parkinson’s disease. PLoS ONE 2014, 9, e93751.
- Bhatwadekar, A.D.; Yan, Y.; Stepps, V.; Hazra, S.; Korah, M.; Bartelmez, S.; Chaqour, B.; Grant, M.B. miR-92a Corrects CD34+ Cell Dysfunction in Diabetes by Modulating Core Circadian Genes Involved in Progenitor Differentiation. Diabetes 2015, 64, 4226–4237.
- Setyowati Karolina, D.; Sepramaniam, S.; Tan, H.Z.; Armugam, A.; Jeyaseelan, K. miR-25 and miR-92a regulate insulin I biosynthesis in rats. RNA Biol. 2013, 10, 1365–1378.
- Peng, T.; Liu, X.; Wang, J.; Liu, Y.; Fu, Z.; Ma, X.; Li, J.; Sun, G.; Ji, Y.; Lu, J.; et al. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2764–2774.
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790.
- Pek, S.L.; Sum, C.F.; Lin, M.X.; Cheng, A.K.; Wong, M.T.; Lim, S.C.; Tavintharan, S. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Mol. Cell. Endocrinol. 2016, 427, 112–123.
- Barbagallo, C.; Mostile, G.; Baglieri, G.; Giunta, F.; Luca, A.; Raciti, L.; Zappia, M.; Purrello, M.; Ragusa, M.; Nicoletti, A. Specific Signatures of Serum miRNAs as Potential Biomarkers to Discriminate Clinically Similar Neurodegenerative and Vascular-Related Diseases. Cell. Mol. Neurobiol. 2020, 40, 531–546.
- de Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980.
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831.
- Frost, R.J.A.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080.
- Martinez, B.; Peplow, P.V. MicroRNAs in Parkinson’s disease and emerging therapeutic targets. Neural. Regen Res. 2017, 12, 1945–1959.
- Wu, J.; Lu, K.; Zhu, M.; Xie, X.; Ding, Y.; Shao, X.; Chen, Y.; Liu, J.; Xu, M.; Xu, Y.; et al. miR-485 suppresses inflammation and proliferation of mesangial cells in an in vitro model of diabetic nephropathy by targeting NOX5. Biochem. Biophys. Res. Commun. 2020, 521, 984–990.
- da Silva, F.C.; Iop, R.D.; Vietta, G.G.; Kair, D.A.; Gutierres Filho, P.J.; de Alvarenga, J.G.; da Silva, R. microRNAs involved in Parkinson’s disease: A systematic review. Mol. Med. Rep. 2016, 14, 4015–4022.
- Jiang, F.; Zong, Y.; Ma, X.; Jiang, C.; Shan, H.; Lin, Y.; Xia, W.; Yin, F.; Wang, N.; Zhou, L.; et al. miR-26a Attenuated Bone-Specific Insulin Resistance and Bone Quality in Diabetic Mice. Mol. Ther. Nucleic Acids 2020, 20, 459–467.
- Xu, H.; Du, X.; Xu, J.; Zhang, Y.; Tian, Y.; Liu, G.; Wang, X.; Ma, M.; Du, W.; Liu, Y.; et al. Pancreatic β cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving β cell function. PLoS Biol. 2020, 18, e3000603.
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J. Clin. Investig. 2015, 125, 2497–2509.
- Oliveira, S.R.; Dionísio, P.A.; Correia Guedes, L.; Gonçalves, N.; Coelho, M.; Rosa, M.M.; Amaral, J.D.; Ferreira, J.J.; Rodrigues, C. Circulating Inflammatory miRNAs Associated with Parkinson’s Disease Pathophysiology. Biomolecules 2020, 10, 945.
- Caggiu, E.; Paulus, K.; Mameli, G.; Arru, G.; Sechi, G.P.; Sechi, L.A. Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. Eneurological. Sci. 2018, 13, 1–4.
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Torkamandi, S.; Saffari, S.; Iranpour, M.; Omrani, M.D. Association of MiR-146a Expression and Type 2 Diabetes Mellitus: A Meta-Analysis. Int. J. Mol. Cell Med. 2017, 6, 156–163.
- García-Jacobo, R.E.; Uresti-Rivera, E.E.; Portales-Pérez, D.P.; González-Amaro, R.; Lara-Ramírez, E.E.; Enciso-Moreno, J.A.; García-Hernández, M.H. Circulating miR-146a, miR-34a and miR-375 in type 2 diabetes patients, pre-diabetic and normal-glycaemic individuals in relation to β-cell function, insulin resistance and metabolic parameters. Clin. Exp. Pharmacol. Physiol. 2019, 46, 1092–1100.
- Mensà, E.; Giuliani, A.; Matacchione, G.; Gurău, F.; Bonfigli, A.R.; Romagnoli, F.; De Luca, M.; Sabbatinelli, J.; Olivieri, F. Circulating miR-146a in healthy aging and type 2 diabetes: Age- and gender-specific trajectories. Mech. Ageing Dev. 2019, 180, 1–10.
- Assmann, T.S.; Recamonde-Mendoza, M.; Puñales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res. Clin. Pract. 2018, 141, 35–46.
- Salunkhe, V.A.; Ofori, J.K.; Gandasi, N.R.; Salö, S.A.; Hansson, S.; Andersson, M.E.; Wendt, A.; Barg, S.; Esguerra, J.; Eliasson, L. MiR-335 overexpression impairs insulin secretion through defective priming of insulin vesicles. Physiol. Rep. 2017, 5.
- Tang, X.W.; Qin, Q.X. miR-335-5p induces insulin resistance and pancreatic islet β-cell secretion in gestational diabetes mellitus mice through VASH1-mediated TGF-β signaling pathway. J. Cell. Physiol. 2019, 234, 6654–6666.
- Khamaneh, A.M.; Alipour, M.R.; Sheikhzadeh Hesari, F.; Ghadiri Soufi, F. A signature of microRNA-155 in the pathogenesis of diabetic complications. J. Physiol. Biochem. 2015, 71, 301–309.
- Akhbari, M.; Khalili, M.; Shahrabi-Farahani, M.; Biglari, A.; Bandarian, F. Expression Level of Circulating Cell Free miR-155 Gene in Serum of Patients with Diabetic Nephropathy. Clin. Lab. 2019, 65, 1493–1499.
- Roser, A.E.; Caldi Gomes, L.; Schünemann, J.; Maass, F.; Lingor, P. Circulating miRNAs as diagnostic biomarkers for Parkinson’s disease. Front. Neurosci. 2018, 12, 625.
- Yildirim, S.S.; Akman, D.; Catalucci, D.; Turan, B. Relationship Between Downregulation of miRNAs and Increase of Oxidative Stress in the Development of Diabetic Cardiac Dysfunction: Junctin as a Target Protein of miR-1. Cell Biochem. Biophys. 2013, 67, 1397–1408.
- de Gonzalo-Calvo, D.; van der Meer, R.W.; Rijzewijk, L.J.; Smit, J.W.; Revuelta-Lopez, E.; Nasarre, L.; Escola-Gil, J.C.; Lamb, H.J.; Llorente-Cortes, V. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci. Rep. 2017, 7, 47.
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019, 66, 226–239.
- Heman-Ackah, S.M.; Hallegger, M.; Rao, M.S.; Wood, M.J. RISC in PD: The impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Front. Mol. Neurosci. 2013, 6, 40.
- Copier, C.U.; León, L.; Fernández, M.; Contador, D.; Calligaris, S.D. Circulating miR-19b and miR-181b are potential biomarkers for diabetic cardiomyopathy. Sci. Rep. 2017, 7, 13514.
- Sun, Y.; Zhou, S.; Shi, Y.; Zhou, Y.; Zhang, Y.; Liu, K.; Zhu, Y.; Han, X. Inhibition of miR-153, an IL-1β-responsive miRNA, prevents beta cell failure and inflammation-associated diabetes. Metabolism 2020, 111, 154335.
- He, J.; Kang, Y.; Lian, C.; Wu, J.; Zhou, H.; Ye, X. Effect of miR-19b on the protective effect of Exendin-4 on islet cells in non-obese diabetic mice. Exp. Ther. Med. 2019, 18, 503–508.
- Mandemakers, W.; Abuhatzira, L.; Xu, H.; Caromile, L.A.; Hébert, S.S.; Snellinx, A.; Morais, V.A.; Matta, S.; Cai, T.; Notkins, A.L.; et al. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2β: Identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia 2013, 56, 1547–1556.
- Tu, C.; Wang, L.; Tao, H.; Gu, L.; Zhu, S.; Chen, X. Expression of miR-409-5p in gestational diabetes mellitus and its relationship with insulin resistance. Exp. Ther. Med. 2020, 20, 3324–3329.
- Ventriglia, G.; Mancarella, F.; Sebastiani, G.; Cook, D.P.; Mallone, R.; Mathieu, C.; Gysemans, C.; Dotta, F. miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2020, 63, 124–136.
- Zhang, Q.; Xiao, X.; Li, M.; Li, W.; Yu, M.; Zhang, H.; Wang, Z.; Xiang, H. Acarbose reduces blood glucose by activating miR-10a-5p and miR-664 in diabetic rats. PLoS ONE 2013, 8, e79697.
- Vasu, S.; Kumano, K.; Darden, C.M.; Rahman, I.; Lawrence, M.C.; Naziruddin, B. MicroRNA Signatures as Future Biomarkers for Diagnosis of Diabetes States. Cells 2019, 8, 1533.
- Zhou, J.; Zhao, Y.; Li, Z.; Zhu, M.; Wang, Z.; Li, Y.; Xu, T.; Feng, D.; Zhang, S.; Tang, F.; et al. miR-103a-3p regulates mitophagy in Parkinson’s disease through Parkin/Ambra1 signaling. Pharmacol. Res. 2020, 160, 105197.
- Serafin, A.; Foco, L.; Zanigni, S.; Blankenburg, H.; Picard, A.; Zanon, A.; Giannini, G.; Pichler, I.; Facheris, M.F.; Cortelli, P.; et al. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology 2015, 84, 645–653.
- Talepoor Ardakani, M.; Rostamian Delavar, M.; Baghi, M.; Nasr-Esfahani, M.H.; Kiani-Esfahani, A.; Ghaedi, K. Upregulation of miR-200a and miR-204 in MPP+-treated differentiated PC12 cells as a model of Parkinson’s disease. Mol. Genet. Genom. Med. 2019, 7, e548.
- Fu, J.; Peng, L.; Tao, T.; Chen, Y.; Li, Z.; Li, J. Regulatory roles of the miR-200 family in neurodegenerative diseases. Biomed. Pharmacother. 2019, 119, 109409.
- Xu, G.; Thielen, L.A.; Chen, J.; Grayson, T.B.; Grimes, T.; Bridges, S.L., Jr.; Tse, H.M.; Smith, B.; Patel, R.; Li, P.; et al. Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E723–E730.
- Jo, S.; Chen, J.; Xu, G.; Grayson, T.B.; Thielen, L.A.; Shalev, A. miR-204 Controls Glucagon-Like Peptide 1 Receptor Expression and Agonist Function. Diabetes 2018, 67, 256–264.
- Gaddam, R.R.; Jacobsen, V.P.; Kim, Y.R.; Gabani, M.; Jacobs, J.S.; Dhuri, K.; Kumar, S.; Kassan, M.; Li, Q.; Bahal, R.; et al. Microbiota-governed microRNA-204 impairs endothelial function and blood pressure decline during inactivity in db/db mice. Sci. Rep. 2020, 10, 10065.
- Su, C.H.; Yang, X.P.; Lou, J.Y. Geniposide reduces alpha-synuclein by blocking microRNA-21/lysosome-associated membrane protein 2A interaction in Parkinson disease models. Brain Res. 2016, 1644, 98–106.
- Zhao, L.; Wang, Z. MicroRNAs: Game changers in the regulation of α-synuclein in Parkinson’s disease. Parkinson’s Dis. 2019, 2019, 1743183.
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18.
- La Sala, L.; Mrakic-Sposta, S.; Micheloni, S.; Prattichizzo, F.; Ceriello, A. Glucose-sensing microRNA-21 disrupts ROS homeostasis and impairs antioxidant responses in cellular glucose variability. Cardiovasc. Diabetol. 2018, 17, 105.
- Chen, Q.; Qiu, F.; Zhou, K.; Matlock, H.G.; Takahashi, Y.; Rajala, R.; Yang, Y.; Moran, E.; Ma, J.X. Pathogenic Role of microRNA-21 in Diabetic Retinopathy Through Downregulation of PPAR alpha. Diabetes 2017, 66, 1671–1682.
- Zang, J.; Maxwell, A.P.; Simpson, D.A.; McKay, G.J. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci. Rep. 2019, 9, 10900.
- Grieco, G.E.; Cataldo, D.; Ceccarelli, E.; Nigi, L.; Catalano, G.; Brusco, N.; Mancarella, F.; Ventriglia, G.; Fondelli, C.; Guarino, E.; et al. Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA 2018, 4, 37.
- Demirsoy, İ.H.; Ertural, D.Y.; Balci, Ş.; Çınkır, Ü.; Sezer, K.; Tamer, L.; Aras, N. Profiles of Circulating MiRNAs Following Metformin Treatment in Patients with Type 2 Diabetes. J. Med. Biochem. 2018, 37, 499–506.
- Mazzeo, A.; Lopatina, T.; Gai, C.; Trento, M.; Porta, M.; Beltramo, E. Functional analysis of miR-21-3p, miR-30b-5p and miR-150-5p shuttled by extracellular vesicles from diabetic subjects reveals their association with diabetic retinopathy. Exp. Eye Res. 2019, 184, 56–63.
- Dong, Y.; Han, L.-L.; Xu, Z.-X.J.M.M. Suppressed microRNA-96 inhibits iNOS expression and dopaminergic neuron apoptosis through inactivating the MAPK signaling pathway by targeting CACNG5 in mice with Parkinson’s disease. Mol. Med. 2018, 24, 61.
- Xie, Y.M.; Chen, Y.H. microRNAs: Emerging Targets Regulating Oxidative Stress in the Models of Parkinson’s Disease. Front. Neurosci. 2016, 10, 298.
- Yang, W.M.; Min, K.H.; Lee, W. Induction of miR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039.
- Jeong, H.J.; Park, S.Y.; Yang, W.M.; Lee, W. The induction of miR-96 by mitochondrial dysfunction causes impaired glycogen synthesis through translational repression of IRS-1 in SK-Hep1 cells. Biochem. Biophys. Res. Commun. 2013, 434, 503–508.
- Behbahanipour, M.; Peymani, M.; Salari, M.; Hashemi, M.S.; Nasr-Esfahani, M.H.; Ghaedi, K. Expression Profiling of Blood microRNAs 885, 361, and 17 in the Patients with the Parkinson’s disease: Integrating Interaction Data to Uncover the Possible Triggering Age-Related Mechanisms. Sci. Rep. 2019, 9, 1–11.
- Su, L.; Wang, C.; Zheng, C.; Wei, H.; Song, X. A meta-analysis of public microarray data identifies biological regulatory networks in Parkinson’s disease. BMC Med. Genom. 2018, 11, 40.
- Shaker, O.G.; Abdelaleem, O.O.; Mahmoud, R.H.; Abdelghaffar, N.K.; Ahmed, T.I.; Said, O.M.; Zaki, O.M. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. Iubmb Life 2019, 71, 310–320.
- Satake, E.; Pezzolesi, M.G.; Md Dom, Z.I.; Smiles, A.M.; Niewczas, M.A.; Krolewski, A.S. Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes. Diabetes 2018, 67, 1013–1023.
- Wang, J.; Zhang, J.; Chen, X.; Yang, Y.; Wang, F.; Li, W.; Awuti, M.; Sun, Y.; Lian, C.; Li, Z.; et al. miR-365 promotes diabetic retinopathy through inhibiting Timp3 and increasing oxidative stress. Exp. Eye Res. 2018, 168, 89–99.
- Grieco, G.E.; Brusco, N.; Licata, G.; Nigi, L.; Formichi, C.; Dotta, F.; Sebastiani, G. Targeting microRNAs as a Therapeutic Strategy to Reduce Oxidative Stress in Diabetes. Int. J. Mol. Sci. 2019, 20, 6358.
- Wang, S.S.; Li, Y.Q.; Liang, Y.Z.; Dong, J.; He, Y.; Zhang, L.; Yan, Y.X. Expression of miR-18a and miR-34c in circulating monocytes associated with vulnerability to type 2 diabetes mellitus and insulin resistance. J. Cell Mol. Med. 2017, 21, 3372–3380.
- Xu, X.H.; Ding, D.F.; Yong, H.J.; Dong, C.L.; You, N.; Ye, X.L.; Pan, M.L.; Ma, J.H.; You, Q.; Lu, Y.B. Resveratrol transcriptionally regulates miRNA-18a-5p expression ameliorating diabetic nephropathy via increasing autophagy. Eur. Rev. Med. Pharm. Sci. 2017, 21, 4952–4965.
- Mononen, N.; Lyytikäinen, L.P.; Seppälä, I.; Mishra, P.P.; Juonala, M.; Waldenberger, M.; Klopp, N.; Illig, T.; Leiviskä, J.; Loo, B.M.; et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Sci. Rep. 2019, 9, 8887.
- Dos Santos, M.; Barreto-Sanz, M.A.; Correia, B.; Bell, R.; Widnall, C.; Perez, L.T.; Berteau, C.; Schulte, C.; Scheller, D.; Berg, D.; et al. miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 2018, 9, 17455.
- Xu, L.; Li, Y.; Yin, L.; Qi, Y.; Sun, H.; Sun, P.; Xu, M.; Tang, Z.; Peng, J. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics 2018, 8, 5593–5609.
- Herrera, B.M.; Lockstone, H.E.; Taylor, J.M.; Wills, Q.F.; Kaisaki, P.J.; Barrett, A.; Camps, C.; Fernandez, C.; Ragoussis, J.; Gauguier, D.; et al. MicroRNA-125a is over-expressed in insulin target tissues in a spontaneous rat model of Type 2 Diabetes. BMC Med. Genom. 2009, 2, 54.
- Fan, Y.; Zhao, X.; Lu, K.; Cheng, G. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res. Bull. 2020, 157, 119–127.
- Yu, C.Y.; Yang, C.Y.; Rui, Z.L. MicroRNA-125b-5p improves pancreatic beta-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci. 2019, 224, 67–75.
- Shen, Y.; Xu, H.; Pan, X.; Wu, W.; Wang, H.; Yan, L.; Zhang, M.; Liu, X.; Xia, S.; Shao, Q. miR-34a and miR-125b are upregulated in peripheral blood mononuclear cells from patients with type 2 diabetes mellitus. Exp. Ther. Med. 2017, 14, 5589–5596.
- Cheung, R.; Pizza, G.; Rolando, D.M.; Chabosseau, P.L.; Nguyen-TU, M.S.; Leclerc, I.; Rutter, G.A.; Martinez-Sanchez, A.I.D.A. 2183-P: miR-125b Is Regulated by Glucose via AMPK and Impairs ß-Cell Function. Am. Diabetes Assoc. 2019, 68, 2183.
- Lamadrid-Romero, M.; Solís, K.H.; Cruz-Reséndiz, M.S.; Pérez, J.E.; Díaz, N.F.; Flores-Herrera, H.; García-López, G.; Perichart, O.; Reyes-Muñoz, E.; Arenas-Huertero, F.; et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci. Res. 2018, 130, 8–22.
- Hoss, A.G.; Labadorf, A.; Beach, T.G.; Latourelle, J.C.; Myers, R.H. RNA profiles in Parkinson’s disease prefrontal cortex. Front. Aging Neurosci. 2016, 8, 36.
- Parrizas, M.; Mundet, X.; Castaño, C.; Canivell, S.; Cos, X.; Brugnara, L.; Giráldez-García, C.; Regidor, E.; Mata-Cases, M.; Franch-Nadal, J.; et al. miR-10b and miR-223-3p in serum microvesicles signal progression from prediabetes to type 2 diabetes. J. Endocrinol. Investig. 2020, 43, 451–459.
- Åkerman, L.; Casas, R.; Ludvigsson, J.; Tavira, B.; Skoglund, C. Serum miRNA levels are related to glucose homeostasis and islet autoantibodies in children with high risk for type 1 diabetes. PLoS ONE 2018, 13, e0191067.
- Kwon, D.; Liew, H.J.M.; Toxicology, C. miRNA profile of neuroprotection mechanism of echinomycin in Parkinson’s disease. Mol. Cell. Toxicol. 2017, 13, 229–238.
- Zhang, H.; Liu, J.; Qu, D.; Wang, L.; Luo, J.Y.; Lau, C.W.; Liu, P.; Gao, Z.; Tipoe, G.L.; Lee, H.; et al. Inhibition of miR-200c Restores Endothelial Function in Diabetic Mice Through Suppression of COX-2. Diabetes 2016, 65, 1196–1207.
- Watts, M.E.; Williams, S.M.; Nithianantharajah, J.; Claudianos, C. Hypoxia-Induced MicroRNA-210 Targets Neurodegenerative Pathways. Noncoding RNA 2018, 4, 10.
- Pordzik, J.; Jakubik, D.; Jarosz-Popek, J.; Wicik, Z.; Eyileten, C.; De Rosa, S.; Indolfi, C.; Siller-Matula, J.M.; Czajka, P.; Postula, M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: Bioinformatic analysis and review. Cardiovasc. Diabetol. 2019, 18, 113.
- Xing, R.X.; Li, L.G.; Liu, X.W.; Tian, B.X.; Cheng, Y. Down regulation of miR-218, miR-124, and miR-144 relates to Parkinson’s disease via activating NF-κB signaling. Kaohsiung J. Med. Sci. 2020, 36, 786–792.
- Kong, Q.; Guo, X.; Guo, Z.; Su, T. Urinary Exosome miR-424 and miR-218 as Biomarkers for Type 1 Diabetes in Children. Clin. Lab. 2019, 65, 937–946.
- Yao, R.; Yao, X.; Liu, R.; Peng, J.; Tian, T. Glucose-induced microRNA-218 suppresses the proliferation and promotes the apoptosis of human retinal pigment epithelium cells by targeting RUNX2. Biosci. Rep. 2019, 39, BSR20192580.
- Lang, H.; Ai, Z.; You, Z.; Wan, Y.; Guo, W.; Xiao, J.; Jin, X. Characterization of miR-218/322-Stxbp1 pathway in the process of insulin secretion. J. Mol. Endocrinol. 2015, 54, 65–73.
- Du, H.; Fu, Z.; He, G.; Wang, Y.; Xia, G.; Fang, M.; Zhang, T. MicroRNA-218 targets adiponectin receptor 2 to regulate adiponectin signaling. Mol. Med. Rep. 2015, 11, 4701–4705.
- Mortuza, R.; Feng, B.; Chakrabarti, S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia 2014, 57, 1037–1046.
- Wang, J.; Pan, Y.; Dai, F.; Wang, F.; Qiu, H.; Huang, X. Serum miR-195-5p is upregulated in gestational diabetes mellitus. J. Clin. Lab. Anal. 2020, 8, e23325.
- Zheng, D.; Ma, J.; Yu, Y.; Li, M.; Ni, R.; Wang, G.; Chen, R.; Li, J.; Fan, G.C.; Lacefield, J.C.; et al. Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia 2015, 58, 1949–1958.
- Wan, S.; Wang, J.; Wang, J.; Wu, J.; Song, J.; Zhang, C.Y.; Zhang, C.; Wang, C.; Wang, J.J. Increased serum miR-7 is a promising biomarker for type 2 diabetes mellitus and its microvascular complications. Diabetes Res. Clin. Pract. 2017, 130, 171–179.
- Wang, Y.; Liu, J.; Liu, C.; Naji, A.; Stoffers, D.A. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes 2013, 62, 887–895.
- Roser, A.E.; Caldi Gomes, L.; Halder, R.; Jain, G.; Maass, F.; Tönges, L.; Tatenhorst, L.; Bähr, M.; Fischer, A.; Lingor, P. miR-182-5p and miR-183-5p Act as GDNF Mimics in Dopaminergic Midbrain Neurons. Mol. Ther. Nucleic Acids 2018, 11, 9–22.
- Zhang, D.; Li, Y.; Yao, X.; Wang, H.; Zhao, L.; Jiang, H.; Yao, X.; Zhang, S.; Ye, C.; Liu, W.; et al. miR-182 Regulates Metabolic Homeostasis by Modulating Glucose Utilization in Muscle. Cell Rep. 2016, 16, 757–768.
- Zhou, J.; Meng, Y.; Tian, S.; Chen, J.; Liu, M.; Zhuo, M.; Zhang, Y.; Du, H.; Wang, X. Comparative MicroRNA Expression Profiles of Cynomolgus Monkeys, Rat, and Human Reveal that miR-182 Is Involved in T2D Pathogenic Processes. J. Diabetes Res. 2014.
- Weale, C.J.; Matshazi, D.M.; Davids, S.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. Circulating miR-30a-5p and miR-182-5p in Prediabetes and Screen-Detected Diabetes Mellitus. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 5037–5047.
- Ba, Q.; Cui, C.; Wen, L.; Feng, S.; Zhou, J.; Yang, K. Schisandrin B shows neuroprotective effect in 6-OHDA-induced Parkinson’s disease via inhibiting the negative modulation of miR-34a on Nrf2 pathway. Biomed. Pharmacother. 2015, 75, 165–172.
- Rostamian Delavar, M.; Baghi, M.; Safaeinejad, Z.; Kiani-Esfahani, A.; Ghaedi, K.; Nasr-Esfahani, M.H. Differential expression of miR-34a, miR-141, and miR-9 in MPP + -treated differentiated PC12 cells as a model of Parkinson’s disease. Gene 2018, 662, 54–65.
- Zhang, X.; Yang, R.; Hu, B.L.; Lu, P.; Zhou, L.L.; He, Z.Y.; Wu, H.M.; Zhu, J.H. Reduced Circulating Levels of miR-433 and miR-133b Are Potential Biomarkers for Parkinson’s Disease. Front. Cell Neurosci. 2017, 11, 170.
- Xie, Y.; Jia, Y.; Cuihua, X.; Hu, F.; Xue, M.; Xue, Y. Urinary Exosomal MicroRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J. Diabetes Res. 2017, 2017, 6978984.
- Chen, Y.; Zheng, J.; Su, L.; Chen, F.; Zhu, R.; Chen, X.; Ye, Q. Increased Salivary microRNAs That Regulate DJ-1 Gene Expression as Potential Markers for Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 210.
- Cui, C.; Ye, X.; Chopp, M.; Venkat, P.; Zacharek, A.; Yan, T.; Ning, R.; Yu, P.; Cui, G.; Chen, J. miR-145 Regulates Diabetes-Bone Marrow Stromal Cell-Induced Neurorestorative Effects in Diabetes Stroke Rats. Stem Cells Transl. Med. 2016, 5, 1656–1667.
- Riches, K.; Alshanwani, A.R.; Warburton, P.; O’Regan, D.J.; Ball, S.G.; Wood, I.C.; Turner, N.A.; Porter, K.E. Elevated expression levels of miR-143/5 in saphenous vein smooth muscle cells from patients with Type 2 diabetes drive persistent changes in phenotype and function. J. Mol. Cell Cardiol. 2014, 74, 240–250.
- Xihua, L.; Shengjie, T.; Weiwei, G.; Matro, E.; Tingting, T.; Lin, L.; Fang, W.; Jiaqiang, Z.; Fenping, Z.; Hong, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2019, 205, 33–43.
- Ghanbari, M.; Darweesh, S.K.; de Looper, H.W.; van Luijn, M.M.; Hofman, A.; Ikram, M.A.; Franco, O.H.; Erkeland, S.J.; Dehghan, A. Genetic Variants in MicroRNAs and Their Binding Sites Are Associated with the Risk of Parkinson Disease. Hum. Mutat. 2016, 37, 292–300.
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.; Rodriguez-Oroz, M.C.; Obeso, J.A.; Cooper, J.M. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell death Dis. 2013, 4, e545.
- Stępień, E.Ł.; Durak-Kozica, M.; Kamińska, A.; Targosz-Korecka, M.; Libera, M.; Tylko, G.; Opalińska, A.; Kapusta, M.; Solnica, B.; Georgescu, A.; et al. Circulating ectosomes: Determination of angiogenic microRNAs in type 2 diabetes. Theranostics 2018, 8, 3874–3890.
- Liang, Y.Z.; Li, J.J.; Xiao, H.B.; He, Y.; Zhang, L.; Yan, Y.X. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: A systematic review and meta-analysis. J. Diabetes 2020, 12, 633–644.
- Zeng, R.; Luo, D.X.; Li, H.P.; Zhang, Q.S.; Lei, S.S.; Chen, J.H. MicroRNA-135b alleviates MPP+-mediated Parkinson’s disease in in vitro model through suppressing FoxO1-induced NLRP3 inflammasome and pyroptosis. J. Clin. Neurosci. 2019, 65, 125–133.
- Gong, Q.; Xie, J.; Liu, Y.; Li, Y.; Su, G. Differentially Expressed MicroRNAs in the Development of Early Diabetic Retinopathy. J. Diabetes Res. 2017, 2017, 4727942.
- Senese, R.; Cioffi, F.; Petito, G.; de Lange, P.; Russo, A.; Goglia, F.; Lanni, A.; Potenza, N. miR-22-3p is involved in gluconeogenic pathway modulated by 3,5-diiodo-L-thyronine (T2). Sci. Rep. 2019, 9, 1–9.
- Kaur, K.; Vig, S.; Srivastava, R.; Mishra, A.; Singh, V.P.; Srivastava, A.K.; Datta, M. Elevated Hepatic miR-22-3p Expression Impairs Gluconeogenesis by Silencing the Wnt-Responsive Transcription Factor Tcf7. Diabetes 2015, 64, 3659–3669.
- Estrella, S.; Garcia-Diaz, D.F.; Codner, E.; Camacho-Guillén, P.; Pérez-Bravo, F. Expression of miR-22 and miR-150 in type 1 diabetes mellitus: Possible relationship with autoimmunity and clinical characteristics. Med. Clin. 2016, 147, 245–247.
- Ye, D.; Zhang, T.; Lou, G.; Xu, W.; Dong, F.; Chen, G.; Liu, Y. Plasma miR-17, miR-20a, miR-20b and miR-122 as potential biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus patients. Life Sci. 2018, 208, 201–207.
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352.
- Santosh, P.S.; Arora, N.; Sarma, P.; Pal-Bhadra, M.; Bhadra, U. Interaction Map and Selection of microRNA Targets in Parkinson’s Disease-Related Genes. J. Biomed. Biotechnol. 2009, 2009, 363145.
- Vallelunga, A.; Iannitti, T.; Dati, G.; Capece, S.; Maugeri, M.; Tocci, E.; Picillo, M.; Volpe, G.; Cozzolino, A.; Squillante, M.; et al. Serum miR-30c-5p is a potential biomarker for multiple system atrophy. Mol. Biol. Rep. 2019, 46, 1661–1666.
- Vallelunga, A.; Ragusa, M.; Di Mauro, S.; Iannitti, T.; Pilleri, M.; Biundo, R.; Weis, L.; Di Pietro, C.; De Iuliis, A.; Nicoletti, A.; et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front. Cell. Neurosci. 2014, 8, 156.
- Irani, S.; Iqbal, J.; Antoni, W.J.; Ijaz, L.; Hussain, M.M. microRNA-30c reduces plasma cholesterol in homozygous familial hypercholesterolemic and type 2 diabetic mouse models. J. Lipid Res. 2018, 59, 144–154.
- Yan, L.N.; Zhang, X.; Xu, F.; Fan, Y.Y.; Ge, B.; Guo, H.; Li, Z.L. Four-microRNA signature for detection of type 2 diabetes. World J. Clin. Cases 2020, 8, 1923–1931.
- Bai, X.; Tang, Y.; Yu, M.; Wu, L.; Liu, F.; Ni, J.; Wang, Z.; Wang, J.; Fei, J.; Wang, W.; et al. Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci. Rep. 2017, 7, 1–7.
- Dooley, J.; Garcia-Perez, J.E.; Sreenivasan, J.; Schlenner, S.M.; Vangoitsenhoven, R.; Papadopoulou, A.S.; Tian, L.; Schonefeldt, S.; Serneels, L.; Deroose, C.; et al. The microRNA-29 Family Dictates the Balance between Homeostatic and Pathological Glucose Handling in Diabetes and Obesity. Diabetes 2016, 65, 53–61.
- Massart, J.; Sjögren, R.; Lundell, L.S.; Mudry, J.M.; Franck, N.; O’Gorman, D.J.; Egan, B.; Zierath, J.R.; Krook, A. Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes 2017, 66, 1807–1818.
- Arshad, A.R.; Sulaiman, S.A.; Saperi, A.A.; Jamal, R.; Mohamed Ibrahim, N.; Abdul Murad, N.A. MicroRNAs and Target Genes As Biomarkers for the Diagnosis of Early Onset of Parkinson Disease. Front. Mol. Neurosci. 2017, 10, 352.
- Massaro, J.D.; Polli, C.D.; Costa E Silva, M.; Alves, C.C.; Passos, G.A.; Sakamoto-Hojo, E.T.; Rodrigues de Holanda Miranda, W.; Bispo Cezar, N.J.; Rassi, D.M.; Crispim, F.; et al. Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol. Cell Endocrinol. 2019, 490, 1–14.
- Leggio, L.; Vivarelli, S.; L’Episcopo, F.; Tirolo, C.; Caniglia, S.; Testa, N.; Marchetti, B.; Iraci, N. microRNAs in Parkinson’s disease: From pathogenesis to novel diagnostic and therapeutic approaches. Int. J. Mol. Sci. 2017, 18, 2698.
- Jie, R.; Zhu, P.; Zhong, J.; Zhang, Y.; Wu, H. LncRNA KCNQ1OT1 affects cell proliferation, apoptosis and fibrosis through regulating miR-18b-5p/SORBS2 axis and NF-ĸB pathway in diabetic nephropathy. Diabetol. Metab. Syndr. 2020, 12, 1–11.
- Al-Kafaji, G.; Al-Mahroos, G.; Alsayed, N.A.; Hasan, Z.A.; Nawaz, S.; Bakhiet, M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol. Med. Rep. 2015, 12, 7485–7490.
- Houshmand-Oeregaard, A.; Schrölkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.; Broholm, C.; Mathiesen, E.R.; Clausen, T.D.; Vaag, A.; et al. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771.
- Jiménez-Lucena, R.; Camargo, A.; Alcalá-Diaz, J.F.; Romero-Baldonado, C.; Luque, R.M.; van Ommen, B.; Delgado-Lista, J.; Ordovás, J.M.; Pérez-Martínez, P.; Rangel-Zúñiga, O.A.; et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: From the CORDIOPREV study. Exp. Mol. Med. 2018, 50, 1–12.
- Schwienbacher, C.; Foco, L.; Picard, A.; Corradi, E.; Serafin, A.; Panzer, J.; Zanigni, S.; Blankenburg, H.; Facheris, M.F.; Giannini, G.; et al. Plasma and White Blood Cells Show Different miRNA Expression Profiles in Parkinson’s Disease. J. Mol. Neurosci. 2017, 62, 244–254.
- Hu, D.; Wang, Y.; Zhang, H.; Kong, D. Identification of miR-9 as a negative factor of insulin secretion from beta cells. J. Physiol. Biochem. 2018, 74, 291–299.
- Zhou, X.G.; Xiang, C.P.; Zheng, X.X. miR-132 serves as a diagnostic biomarker in gestational diabetes mellitus and its regulatory effect on trophoblast cell viability. Diagn. Pathol. 2019, 14, 1–7.
- Mziaut, H.; Henniger, G.; Ganss, K.; Hempel, S.; Wolk, S.; McChord, J.; Chowdhury, K.; Ravassard, P.; Knoch, K.P.; Krautz, C.; et al. MiR-132 controls pancreatic beta cell proliferation and survival through Pten/Akt/Foxo3 signaling. Mol. Metab. 2020, 31, 150–162.
- Blum, A.; Meerson, A.; Rohana, H.; Jabaly, H.; Nahul, N.; Celesh, D.; Romanenko, O.; Tamir, S. MicroRNA-423 may regulate diabetic vasculopathy. Clin. Exp. Med. 2019, 19, 469–477.
- Yang, W.; Wang, J.; Chen, Z.; Chen, J.; Meng, Y.; Chen, L.; Chang, Y.; Geng, B.; Sun, L.; Dou, L.; et al. NFE2 Induces miR-423-5p to Promote Gluconeogenesis and Hyperglycemia by Repressing the Hepatic FAM3A-ATP-Akt Pathway. Diabetes 2017, 66, 1819–1832.
- Flowers, E.; Aouizerat, B.E.; Abbasi, F.; Lamendola, C.; Grove, K.M.; Fukuoka, Y.; Reaven, G.M. Circulating microRNA-320a and microRNA-486 predict thiazolidinedione response: Moving towards precision health for diabetes prevention. Metab. Clin. Exp. 2015, 64, 1051–1059.
- Regmi, A.; Liu, G.; Zhong, X.; Hu, S.; Ma, R.; Gou, L.; Zafar, M.I.; Chen, L. Evaluation of Serum microRNAs in Patients with Diabetic Kidney Disease: A Nested Case-Controlled Study and Bioinformatics Analysis. Med. Sci. Monit. 2019, 25, 1699–1708.
- Collares, C.V.; Evangelista, A.F.; Xavier, D.J.; Rassi, D.M.; Arns, T.; Foss-Freitas, M.C.; Foss, M.C.; Puthier, D.; Sakamoto-Hojo, E.T.; Passos, G.A.; et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res. Notes 2013, 6, 491.
- Catanzaro, G.; Besharat, Z.M.; Chiacchiarini, M.; Abballe, L.; Sabato, C.; Vacca, A.; Borgiani, P.; Dotta, F.; Tesauro, M.; Po, A.; et al. Circulating MicroRNAs in Elderly Type 2 Diabetic Patients. Int. J. Endocrinol. 2018, 2018, 6872635.
- Xiao, F.; Li, L.; Fu, J.S.; Hu, Y.X.; Luo, R. Regulation of the miR-19b-mediated SOCS6-JAK2/STAT3 pathway by lncRNA MEG3 is involved in high glucose-induced apoptosis in hRMECs. Biosci. Rep. 2020, 40.





