
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alvaro Ortega | + 1844 word(s) | 1844 | 2021-04-22 08:10:24 | | | |
| 2 | Vivi Li | Meta information modification | 1844 | 2021-04-23 03:20:18 | | |
Video Upload Options
Bacteria possess a large number of signal transduction systems that sense and respond to different environmental cues. Most frequently these are transcriptional regulators, two-component systems and chemosensory pathways. A major bottleneck in the field of signal transduction is the lack of information on signal molecules that modulate the activity of the large majority of these systems.
1. Introduction
Bacteria have evolved an array of different signal transduction mechanisms that are able to sense and respond to a wide range of environmental cues and signal molecules. These systems are generally transcriptional regulators, two-component systems (TCS) and chemosensory pathways [1][2][3] (Figure 1).
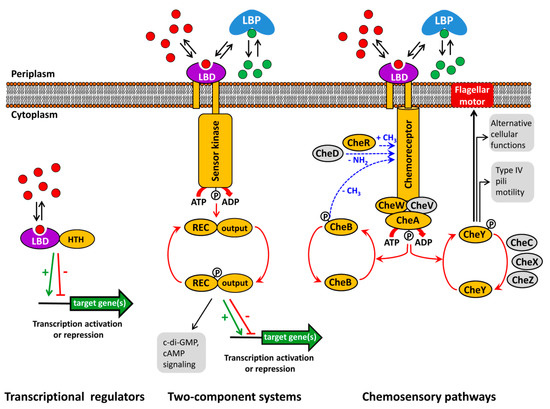
Figure 1. Major bacterial signal transduction systems. LBD: ligand binding domain; HTH: helix-turn-helix motif containing DNA binding domain; LBP: ligand binding protein; REC: receiver domain. The core proteins of chemosensory pathways, present in most pathways, are shaded in yellow, whereas auxiliary proteins, present only in some pathways, are shown in grey. red and green dots: signal molecules; red arrows represent phosphorylation/dephosphorylation events, blue arrows represent methylation/demethylation/deamidation of amino acids; black arrows indicate the pathway output, the green arrow and red T-bar indicate transcriptional activation or inhibition, respectively.
Typically, the capacity of transcriptional regulators to regulate promoter activity is modulated by the recognition of signal molecules. Alternatively, TCS function is based on the signal mediated modulation of the sensor kinase activity that in turn modulates transphosphorylation kinetics to the response regulator. As in the case of transcriptional regulators, the majority of bacterial TCSs appear to be involved in transcriptional regulation [4][5]. Chemosensory pathways can be understood as sophisticated versions of TCSs where the stimulus is received by chemoreceptors that in turn modulate the activity of the CheA histidine kinase leading to alterations in CheY phosphorylation. The majority of chemosensory pathways mediate chemotaxis, whereas others carry out alternative cellular functions or are associated with type IV pili based motility [2][6].
There is an enormous diversity in the domain organization and topology of signal transduction systems [1][3][6][7][8]. This diversity is also reflected in a variety of different mechanisms that regulate receptor activity, such as stimulus mediated alterations in the transmembrane regions of sensor kinases and chemoreceptors [9][10], signal sensing by the cytoplasmic autokinase domain of histidine kinases [11], chemoreceptor activation by proteolysis [12] or via the phosphotransferase system [13].
However, the canonical mode of their activation consists in the binding of signal molecules to the ligand binding domains (LBD) that are present in all three major signal transduction systems (Figure 1). Thus, signal binding to many transcriptional regulators modulates their affinity for promoter regions [14]. In contrast, ligand binding to the LBD of sensor kinases [15] and chemoreceptors [16] was shown to cause piston-like movements of transmembrane regions that ultimately cause changes in autokinase activity. This appears to be a general mechanism that applies to sensor kinases and chemoreceptors, since, firstly, chimeric receptors comprising different types of chemoreceptor LBDs with the cytosolic fragment of the Tar chemoreceptor were functional [17][18] and, secondly, a number of functional chemoreceptor/sensor kinase chimeras were produced [19][20].
The majority of bacterial sensor proteins are of unknown function. For other systems, a function has been identified mainly through the phenotypic characterization of mutant strains. However, in a significant number of cases information on the stimulus that modulates the activity of a given system is lacking. This lack of knowledge on signals recognized by sensor proteins represents a major bottleneck in signal transduction research [21]. This can be best illustrated by the three sensor kinases GacS, LadS and RetS. Although these proteins play a central role in the virulence of the human pathogen Pseudomonas aeruginosa [22], the signals that stimulate these three proteins have so far not been identified.
Sensor protein function is closely related to the signals recognized by the corresponding LBD. However, LBDs show important sequence diversity and, as a result, the ligand specificity of a given LBD is frequently not reflected in overall LBD sequence homology [23], which in turn hampers functional annotation by extrapolation from homologous systems. This limitation makes experimental approaches for the functional annotation of bacterial sensor proteins essential. In this article we will discuss progress made over mainly the last 10 years in experimental approaches that resulted in the functional annotation of a significant number of bacterial sensor proteins.
2. Functional Annotation Using Genetic Approaches
Insight into the function of genes and proteins can be gained by the phenotypic analysis of bacterial chemoreceptor mutants. Using this approach the function of many chemoreceptors has been identified. As representative examples we would like to cite here the identification of chemoreceptors for naphthalene [24], cyclic carboxylic acids [25], cytosine [26], inorganic phosphate [27] or boric acid [28]. However, this strategy has several limitations that we illustrate here.
2.1. Multiple Receptors
Chemotactic bacteria possess on average 14 chemoreceptor genes [29] and in some case up to 80 genes were detected [30]. There are a number of reports showing that some species possess multiple receptors that respond to the same ligand. In these cases the loss of activity caused by the mutation of a single chemoreceptor gene may be compensated by additional receptors and the analysis of chemoreceptor single mutants may not lead to a functional annotation. For example, wild type Comamonas testosteroni and a mutant defective in the chemoreceptor Mcp2983 had indistinguishable chemotaxis to a series of organic acids and organic compounds. However, the complementation of a chemotaxis free mutant, in which all 22 chemoreceptor genes were deleted, with the mcp2983 gene resulted in the recovery of wild-type like chemotaxis to a number of chemoeffectors [31]. The authors conclude that the genome of C. testosteroni encodes additional chemoreceptors that compensate the deletion of the mcp2983 gene. Another example is Ralstonia pseudosolanaceum that was identified to contain at least two chemoreceptors for citrate. In analogy to the above study, single mutants in each of the two receptors did not alter citrate chemotaxis and a reduction was only observed in the double mutant [32]. Another recent study revealed that chemotaxis to histamine in P. aeruginosa is mediated by the combined action of three chemoreceptors, TlpQ, PctA and PctC. TlpQ binds histamine with very high affinity and a mutant in tlpQ was only defective in histamine chemotaxis at low concentrations of the chemoattractant [33]. Also, the recent analysis of the chemoreceptor repertoire of Bacillus amyloliquefaciens identified multiple receptors that responded to a same amino acid, sugar or organic acid [34]. Further examples are multiple amino acid receptors in P. aeruginosa [35][36], P. fluorescens [37] and Sinorhizobium meliloti [38][39] as well as several chemoreceptors in P. fluorescens Pf0-1 or P. putida KT2440 that respond to Krebs cycle intermediates [40][41][42][43].
2.2. Chemotaxis Is Induced or Repressed by the Cognate Ligands
There is also evidence that some chemoeffectors either induce [25] or repress [27][44] the chemotaxis phenotype. For example, P. aeruginosa does not show any chemotaxis towards inorganic phosphate (Pi) under standard culture conditions in which cells are grown in media containing significant amounts of Pi [27][44]. However, very strong Pi chemotaxis was observed under Pi limiting conditions, which is due to the fact that Pi represses the expression of its cognate chemoreceptors [27][45].
2.3. Energy Taxis May Mask Chemotaxis
Tactic movements can be due to chemotaxis, typically characterised by the recognition of the chemoeffector by a chemoreceptor in the extracytosolic space, or energy taxis, which is based on sensing molecular consequences that occur as a result of chemoeffector metabolization [46]. However, in a number of cases chemo- and energy taxis to a given compound occur simultaneously [47][48]. For example, malate energy taxis in P. aeruginosa was found to dominate and mask malate chemotaxis to an extent that chemotaxis became only visible in a mutant defective in the energy taxis chemoreceptor [48]. Therefore, as concluded before, the phenotypic characterization of chemoreceptor single mutants may not result in identifying the function of these receptors.
3. Three-Dimensional Structural Information Provides Clues on Ligands Recognized by Homologous Sensor Proteins
Ligand binding domains are characterized by a high degree of sequence divergence and frequently the overall sequence similarity between homologous proteins is not reflected in a similarity of ligands recognized [23]. However, three-dimensional structural information is now an invaluable tool to get initial information on ligands recognized. We would like to illustrate this issue on the example of CACHE domains that exist either in a mono-modular (sCACHE) or bi-modular (dCACHE) configuration [49]. CACHE domains are the most abundant sensor domains in both, histidine kinases [50] and chemoreceptors [7].
As significant number of structures deposited in the Protein Data Bank (pdb) are the result of structural genomics initiatives and represent a source of information on the function of homologous proteins. In the framework of structural genomic projects on Anaeromyxobacter dehalogenans and Vibrio parahaemolyticus, structures of sCACHE domains have been deposited at pdb (pdb ID 4K08 and 4EXO, respectively). In both cases, their binding pocket was occupied by a ligand, namely acetate in 4K08 and pyruvate in 4EXO. In the case of the 4K08 structure, acetate was a component of the crystallization buffer. Since pyruvate was absent from the crystallization buffer of the 4EXO structure, the ligand must have been co-purified with the protein. P. putida KT2440 has a single chemoreceptor with a sCACHE domain (PP_2861), which shares only 22% sequence identity with the 4EXO protein. The above observations have directed our research and we have tested whether the LBD of PP_2861 also bound acetate and pyruvate. We were able to show that this was the case and have identified with propionate and L-lactate two additional ligands. The corresponding chemoreceptor was renamed McpP and was found to mediate chemoattraction to these four ligands [51].
Similarly, structural information was also useful to study dCACHE domain. The first structure of a dCACHE domain deposited into pdb was entry with ID 3C8C; solved in a structural genomics project of Vibrio cholerae. In analogy to the case above, the binding pocket of this structure was occupied with a ligand, l-Ala, and since l-Ala was not present in the crystallization buffer, it must have been co-purified with the protein suggesting that it is a physiological ligand. This information has been useful to identify other amino acid sensing chemoreceptors and it was established that dCACHE containing chemoreceptors form the primary family of amino acid responsive chemoreceptors [18].
There are different sub-families within dCACHE domains, namely those that bind C4-dicarboxylic acids like succinate and malate [52][53] or those that bind different amines such as amino acids, GABA, polyamines, taurine, purines or quaternary [23][33][36][39][54][55][56][57][58][59]. Structural information has now provided the molecular detail of C4-dicarboxylic acid [52][53] and amine recognition at different dCACHE domains [33][57][58][59] (Figure 2). In all cases the ligand is bound to the membrane distal module. The comparison of amine recognition patterns in structures from different species (P. aeruginosa, P. putida, V. cholerae, Campylobacter jejuni), and in complex with different ligands (putrescine, histamine, taurine, amino acids) identifies the consensus motif Y-x-d-x(n)-d that coordinates the amino group of ligands through a hydrogen bonding network. Therefore, the sequence analysis of dCACHE domains of unknown function for the presence of this motif can provide first clues on the ligand recognized.
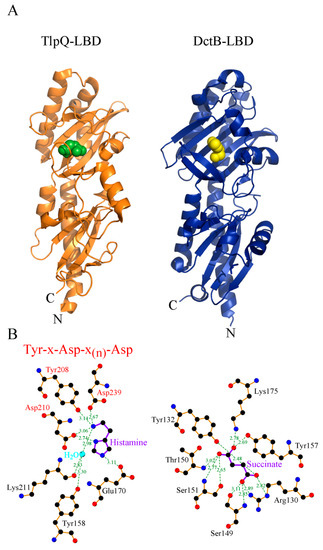
Figure 2. Three-dimensional structure and mode of ligand recognition at dCACHE domains. (A) Ribbon diagram of the LBD of the TlpQ chemoreceptor of P. aeruginosa (orange) in complex with histamine (green, pdb ID 6fu4) and LBD of the DctB sensor kinase of V. cholerae (blue) in complex with succinate (yellow, pdb ID 3by9). (B) Amino acids involved in ligand recognition. The sequence motif involved in amine recognition is shown in red above the histamine plot. Figure generated using Ligplot [60].
References
- Ulrich, L.E.; Koonin, E.V.; Zhulin, I.B. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005, 13, 52–56.
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50.
- Jacob-Dubuisson, F.; Mechaly, A.; Betton, J.M.; Antoine, R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018, 16, 585–593.
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124.
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular Mechanisms of Two-Component Signal Transduction. J. Mol. Biol. 2016, 428, 3752–3775.
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 13, 150–159.
- Ortega, A.; Zhulin, I.B.; Krell, T. Sensory Repertoire of Bacterial Chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81.
- Galperin, M.Y. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 2006, 188, 4169–4182.
- Pham, H.T.; Parkinson, J.S. Phenol sensing by Escherichia coli chemoreceptors: A nonclassical mechanism. J. Bacteriol. 2011, 193, 6597–6604.
- Yuan, J.; Jin, F.; Glatter, T.; Sourjik, V. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. USA 2017, 114, 10792–10798.
- Wang, L.C.; Morgan, L.K.; Godakumbura, P.; Kenney, L.J.; Anand, G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012, 31, 2648–2659.
- Petrova, O.E.; Sauer, K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. Natl. Acad. Sci. USA 2012, 109, 16690–16695.
- Somavanshi, R.; Ghosh, B.; Sourjik, V. Sugar Influx Sensing by the Phosphotransferase System of Escherichia coli. PLoS Biol. 2016, 14, e2000074.
- Balleza, E.; Lopez-Bojorquez, L.N.; Martinez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chavez, I.; Balderas-Martinez, Y.I.; Encarnacion, S.; Collado-Vides, J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2009, 33, 133–151.
- Gushchin, I.; Melnikov, I.; Polovinkin, V.; Ishchenko, A.; Yuzhakova, A.; Buslaev, P.; Bourenkov, G.; Grudinin, S.; Round, E.; Balandin, T.; et al. Mechanism of transmembrane signaling by sensor histidine kinases. Science 2017, 356, eaah6345.
- Ottemann, K.M.; Xiao, W.; Shin, Y.K.; Koshland, D.E., Jr. A piston model for transmembrane signaling of the aspartate receptor. Science 1999, 285, 1751–1754.
- Bi, S.; Pollard, A.M.; Yang, Y.; Jin, F.; Sourjik, V. Engineering Hybrid Chemotaxis Receptors in Bacteria. ACS Synth. Biol. 2016, 5, 989–1001.
- Reyes-Darias, J.A.; Yang, Y.; Sourjik, V.; Krell, T. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol. Microbiol. 2015, 96, 513–525.
- Yoshida, T.; Phadtare, S.; Inouye, M. The design and development of Tar-EnvZ chimeric receptors. Methods Enzymol. 2007, 423, 166–183.
- Ward, S.M.; Delgado, A.; Gunsalus, R.P.; Manson, M.D. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 2002, 44, 709–719.
- Krell, T. Tackling the bottleneck in bacterial signal transduction research: High-throughput identification of signal molecules. Mol. Microbiol. 2015, 96, 685–688.
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364.
- Reyes-Darias, J.A.; Garcia, V.; Rico-Jimenez, M.; Corral-Lugo, A.; Lesouhaitier, O.; Juarez-Hernandez, D.; Yang, Y.; Bi, S.; Feuilloley, M.; Munoz-Rojas, J.; et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 2015, 97, 488–501.
- Grimm, A.C.; Harwood, C.S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 1999, 181, 3310–3316.
- Luu, R.A.; Kootstra, J.D.; Nesteryuk, V.; Brunton, C.N.; Parales, J.V.; Ditty, J.L.; Parales, R.E. Integration of chemotaxis, transport and catabolism in Pseudomonas putida and identification of the aromatic acid chemoreceptor PcaY. Mol. Microbiol. 2015, 96, 134–147.
- Liu, X.; Wood, P.L.; Parales, J.V.; Parales, R.E. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 2009, 191, 2909–2916.
- Wu, H.; Kato, J.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 3400–3404.
- Hida, A.; Oku, S.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of boric acid as a novel chemoattractant and elucidation of its chemoreceptor in Ralstonia pseudosolanacearum Ps29. Sci. Rep. 2017, 7, 8609.
- Lacal, J.; Garcia-Fontana, C.; Munoz-Martinez, F.; Ramos, J.L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884.
- Alexandre, G.; Greer-Phillips, S.; Zhulin, I.B. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 2004, 28, 113–126.
- Ni, B.; Huang, Z.; Wu, Y.F.; Fan, Z.; Jiang, C.Y.; Liu, S.J. A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cis-aconitate and triggers chemotaxis towards diverse organic compounds. Appl. Microbiol. Biotechnol. 2015, 99, 2773–2781.
- Hida, A.; Tajima, T.; Kato, J. Two citrate chemoreceptors involved in chemotaxis to citrate and/or citrate-metal complexes in Ralstonia pseudosolanacearum. J. Biosci. Bioeng. 2018.
- Corral-Lugo, A.; Matilla, M.A.; Martín-Mora, D.; Silva Jiménez, H.; Mesa Torres, N.; Kato, J.; Hida, A.; Oku, S.; Conejero-Muriel, M.; Gavira, J.A.; et al. High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. Mbio 2018, 9, e01894-18.
- Feng, H.; Zhang, N.; Du, W.; Zhang, H.; Liu, Y.; Fu, R.; Shao, J.; Zhang, G.; Shen, Q.R.; Zhang, R. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant Microbe Interact. 2018.
- Taguchi, K.; Fukutomi, H.; Kuroda, A.; Kato, J.; Ohtake, H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 1997, 143, 3223–3229.
- Rico-Jimenez, M.; Munoz-Martinez, F.; Garcia-Fontana, C.; Fernandez, M.; Morel, B.; Ortega, A.; Ramos, J.L.; Krell, T. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol. Microbiol. 2013, 88, 1230–1243.
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 2012, 27, 462–469.
- Webb, B.A.; Hildreth, S.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 2014, 80, 3404–3415.
- Webb, B.A.; Karl Compton, K.; Castaneda Saldana, R.; Arapov, T.D.; Keith Ray, W.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol. Microbiol. 2017, 103, 333–346.
- Oku, S.; Komatsu, A.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of Pseudomonas fluorescens Chemotaxis Sensory Proteins for Malate, Succinate, and Fumarate, and Their Involvement in Root Colonization. Microbes Environ. 2014, 29, 413–419.
- Pineda-Molina, E.; Reyes-Darias, J.A.; Lacal, J.; Ramos, J.L.; Garcia-Ruiz, J.M.; Gavira, J.A.; Krell, T. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc. Natl. Acad. Sci. USA 2012, 109, 18926–18931.
- Martin-Mora, D.; Reyes-Darias, J.A.; Ortega, A.; Corral-Lugo, A.; Matilla, M.A.; Krell, T. McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 2016, 18, 3284–3295.
- Lacal, J.; Garcia-Fontana, C.; Callejo-Garcia, C.; Ramos, J.L.; Krell, T. Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor. J. Mol. Recognit. 2011, 24, 378–385.
- Rico-Jimenez, M.; Reyes-Darias, J.A.; Ortega, A.; Diez Pena, A.I.; Morel, B.; Krell, T. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 28967.
- Bains, M.; Fernandez, L.; Hancock, R.E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 6762–6768.
- Alexandre, G. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 2010, 156, 2283–2293.
- Rabinovitch-Deere, C.A.; Parales, R.E. Three types of taxis used in the response of Acidovorax sp. strain JS42 to 2-nitrotoluene. Appl. Environ. Microbiol. 2012, 78, 2306–2315.
- Alvarez-Ortega, C.; Harwood, C.S. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl. Environ. Microbiol. 2007, 73, 7793–7795.
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016, 12, e1004862.
- Zhang, Z.; Hendrickson, W.A. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 2010, 400, 335–353.
- Garcia, V.; Reyes-Darias, J.A.; Martin-Mora, D.; Morel, B.; Matilla, M.A.; Krell, T. Identification of a Chemoreceptor for C2 and C3 Carboxylic Acids. Appl. Environ. Microbiol. 2015, 81, 5449–5457.
- Cheung, J.; Hendrickson, W.A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265.
- Zhou, Y.F.; Nan, B.; Nan, J.; Ma, Q.; Panjikar, S.; Liang, Y.H.; Wang, Y.; Su, X.D. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008, 383, 49–61.
- Fernandez, M.; Morel, B.; Corral-Lugo, A.; Krell, T. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol. Microbiol. 2016, 99, 34–42.
- Corral-Lugo, A.; de la Torre, J.; Matilla, M.A.; Fernandez, M.; Morel, B.; Espinosa-Urgel, M.; Krell, T. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ. Microbiol. 2016, 18, 3355–3372.
- Matilla, M.A.; Krell, T. Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 2017, 45, 8–14.
- Gavira, J.A.; Ortega, A.; Martin-Mora, D.; Conejero-Muriel, M.T.; Corral-Lugo, A.; Morel, B.; Matilla, M.A.; Krell, T. Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida. J. Mol. Biol. 2018, 430, 1950–1963.
- Nishiyama, S.; Takahashi, Y.; Yamamoto, K.; Suzuki, D.; Itoh, Y.; Sumita, K.; Uchida, Y.; Homma, M.; Imada, K.; Kawagishi, I. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci. Rep. 2016, 6, 20866.
- Liu, Y.C.; Machuca, M.A.; Beckham, S.A.; Gunzburg, M.J.; Roujeinikova, A. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2127–2136.
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134.




