
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Surbhi Sood | + 1135 word(s) | 1135 | 2021-03-29 10:56:11 | | | |
| 2 | Rita Xu | Meta information modification | 1135 | 2021-04-14 08:08:03 | | |
Video Upload Options
In higher-order organisms, the genome is assembled into a protein-dense structure called chromatin. Chromatin is spatially organized in the nucleus through hierarchical folding, which is tightly regulated both in cycling cells and quiescent cells. Assembly and folding are not one-time events in a cell’s lifetime; rather, they are subject to dynamic shifts to allow changes in transcription, DNA replication, or DNA damage repair. Chromatin is regulated at many levels and recent tools have permitted the elucidation of specific factors involved in the maintenance and regulation of the three-dimensional (3D) genome organization.
1. Introduction
1.1. “Control Tower” of the Cell: Chromatin
From the zygote to fully differentiated adult cells, almost every cell in humans contains the exact same DNA content. However, functionally and phenotypically, a dendritic cell and a neuron are different, and their transcriptional response to external cues are different. Basal and inducible gene expression is regulated in part by the way DNA is organized in the nucleus. In higher-order eukaryotic systems, DNA is packed into a protein-dense structure called chromatin, the basic unit of which is called a nucleosome. Nucleosomes consist of an octamer of four types of basic proteins called histones (two copies of each H2A, H2B, H3, and H4); and the ~146 bp of DNA wrapped around this octamer. Another histone protein, H1 is not a part of the nucleosome; however, it plays key roles in connecting nucleosomes and the formation of 3D chromatin structure. Further packing into denser structures, nucleosomes form chromatin fibrils and eventually a 3D organization within the nucleus. It was initially thought that genome packaging is utilized primarily as a way to fit the 2 meters of DNA into the limited space of the nucleus and physically protect it from damage. With advances in molecular biology and molecular genetics, the concept that chromatin is passive and protective has been replaced with the more prominent concept that chromatin contributes to dynamic gene regulation.
1.2. Gene Expression Regulation at the Level of Chromatin Organization
Chromatin is a dynamic structure that responds to extrinsic and intrinsic cues by facilitating the transcription of appropriate genes. Chromatin can be altered by various processes [1], including DNA methylation, non-coding RNAs, histone variants, histone post-translational modifications, and chromatin remodeling [2][3] (Figure 1). These processes are interrelated; the crosstalk among these various factors coordinate the regulation of the chromatin structure. In basic terms, the probability of genic and regulatory DNA interacting with the transcriptional machinery determines whether a gene is expressed or not. When a chromatin region is densely packed into heterochromatin, the DNA is generally not accessible to the transcription machinery; therefore, it will have little or no transcription. In contrast, when chromatin is more loosely packed into euchromatin, the DNA is accessible to transcription machinery, leading to higher transcription. In normal cells, certain chromatin regions do not change their packing density. For instance, constitutive heterochromatin regions comprise centromeric or telomeric sequences or mobile elements, which are kept silent in most of the normal cell types [4], while housekeeping genes, the genes that are essential for basic cellular functions, tend to be constitutively euchromatic. At other loci, the local chromatin state at specific loci can switch between euchromatin and heterochromatin leading to induction or repression of gene expression.
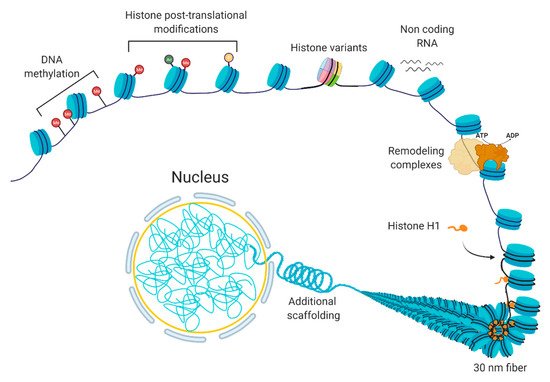
Figure 1. The composition of chromatin. The basic unit consists of DNA and histones, which can be modified or replaced with variants. Additional levels of a structural organization rely on chromatin remodeling, ncRNA expression, and scaffolding proteins, such as histone H1.
2. 3D Chromatin Organization
The existence of organelles facilitates specialized functions within the cell, much like organs within the body. The compartmentalization within organelles is further used to regulate the local concentrations of proteins, thereby coordinating the interaction between enzymes and their substrates. In the nucleus, the spatial organization of the genome controls the local concentration of transcriptional regulators at genes, which may be equally, if not more, critical for localization than specificity for specific genomic elements.
The nucleus is organized into large compartments that can be visualized using microscopy, including the nucleolus, speckles, lamina-associated domains, polycomb bodies, and condensates [5][6][7][8]. Advances in next-generation sequencing technologies have confirmed these compartmental associations and further revealed that chromosomes tend to cluster into transcriptionally active compartments (“A” compartments) and transcriptionally inactive compartments (“B” compartments) (Figure 2) [9]. Within these compartments are megabase-long topologically associating domains (TADs) that remain mostly consistent across cell types, and can even be conserved between species [10]. Within TADs, genomic regions interact with one another at a higher frequency than with regions outside the TAD, and the maintenance of TAD boundaries is important for restricting transcription to within the boundary. The weakening of TAD boundaries results in pathological chromatin contacts that disrupt gene expression, and this has been associated with diseases such as developmental disorders [11], cancer [12], and other genetic diseases [13]. TADs contain additional levels of compartmentalization in the form of sub-TADs and loops, such as ones that link distal enhancers to promoters and recruit large complexes of proteins such as the mediator complex, preinitiation complex (PIC), and other cell-type specific transcription factors, to transcription start sites [14]. Unlike TADs, enhancer-promoter loops vary significantly between cell types [15], although they are restricted to interactions within TAD boundaries (Figure 2).
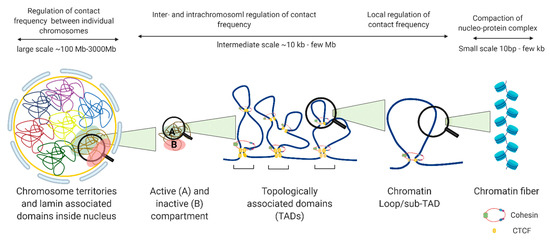
Figure 2. Levels of nuclear genome organization in interphase cells.
There are many proteins involved in directing genome organization, such as lamins, which tether the genome to nuclear membranes [16], and polycomb group proteins, which promote the formation of repressive condensates [17][18][19]; however, CTCF and cohesin are the major architectural factors that define both TAD boundaries and sub-TAD loops [10]. CTCF is a ubiquitously expressed zinc finger protein that acts as an architectural protein by binding at a subset of CCCTC consensus sites across the genome [20]. The majority of long-range chromatin interactions contain consensus sequences for CTCF binding which are pointed toward each other in convergent orientation to allow for homodimerization of CTCF [21][22]. Multiple CTCF motifs are enriched at the boundaries of TADs, which restricts the access of cis-acting elements to regions within the TAD boundaries. It performs this function by cooperating with cohesin, a ring-shaped protein complex that physically restrains chromatin into loops at sites where CTCF homodimers are bound [23]. According to the loop extrusion model, a well-accepted model that explains chromatin folding by architectural proteins cohesin and CTCF, the cohesin complex binds the genome and moves along the DNA, forming a growing knot/loop until it reaches CTCF homodimers bound at convergent CTCF motifs [22][24][25]. Several aspects of this process require repositioning of nucleosomes, such as CTCF binding, cohesin loading, cohesin translocation, and cohesin/CTCF eviction (Figure 3). In this review, we focus on recent genome-wide and mechanistic work investigating the roles for chromatin remodeling complexes in the regulation of CTCF/cohesin-mediated chromatin interactions.
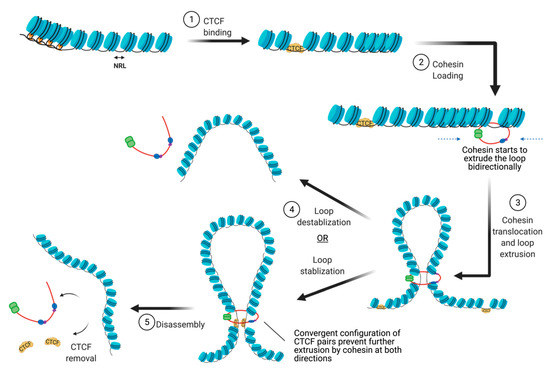
Figure 3. The steps involved in the establishment of CTCF/cohesin mediated looping interactions. (abb: NRL: Nucleosome repeat length).
References
- Dulac, C. Brain function and chromatin plasticity. Nat. Cell Biol. 2010, 465, 728–735.
- Talbert, P.B.; Meers, M.P.; Henikoff, S. Old cogs, new tricks: The evolution of gene expression in a chromatin context. Nat. Rev. Genet. 2019, 20, 283–297.
- Chromatin in Eukaryotic Regulation; NPG Educ.: Cambridge, MA, USA, 2010.
- Ma, Y.; Jacobs, S.B.; Jackson-Grusby, L.; Mastrangelo, M.-A.; Torres-Betancourt, J.A.; Jaenisch, R.; Rasmussen, T.P. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J. Cell Sci. 2005, 118, 1607–1616.
- Fox, A.H.; Lam, Y.W.; Leung, A.K.L.; Lyon, C.E.; Andersen, J.; Mann, M. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25.
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; De Klein, A.; Wessels, L.F.A.; De Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nat. Cell Biol. 2008, 453, 948–951.
- Van Koningsbruggen, S.; Gierliński, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 2010, 21, 3735–3748.
- Pirrotta, V.; Li, H.-B. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012, 22, 101–109.
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293.
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380.
- Lupiáñez, D.G.; Spielmann, M.; Mundlos, S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016, 32, 225–237.
- Akdemir, K.C.; Le, V.T.; Chandran, S.; Li, Y.; Verhaak, R.G.; Beroukhim, R.; Campbell, P.J.; Chin, L.; Dixon, J.R.; Futreal, P.A.; et al. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 2020, 52, 294–305.
- Sun, J.H.; Zhou, L.; Emerson, D.J.; Phyo, S.A.; Titus, K.R.; Gong, W.; Gilgenast, T.G.; Beagan, J.A.; Davidson, B.L.; Tassone, F.; et al. Disease-Associated Short Tandem Repeats Co-localize with Chromatin Domain Boundaries. Cell 2018, 175, 224–238.e15.
- Liu, Z.; Merkurjev, D.; Yang, F.; Li, W.; Oh, S.; Friedman, M.J.; Song, X.; Zhang, F.; Ma, Q.; Ohgi, K.A.; et al. Enhancer Activation Requires trans-Recruitment of a Mega Transcription Factor Complex. Cell 2014, 159, 358–373.
- Dixon, J.R.; Jung, I.; Selvaraj, S.; Shen, Y.; Antosiewicz-Bourget, J.E.; Lee, A.Y. Chromatin architecture reorganization during stem cell differentiation. Nature 2015, 518, 331–336.
- Dittmer, T.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 1–4.
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 2012, 148, 458–472.
- Hou, C.; Li, L.; Qin, Z.S.; Corces, V.G. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol. Cell 2012, 48, 471–484.
- Wani, A.H.; Boettiger, A.N.; Schorderet, P.; Ergun, A.; Münger, C.; Sadreyev, R.I.; Zhuang, X.; Kingston, R.E.; Francis, N.J. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 2016, 7, 10291.
- Farrell, C.M.; West, A.G.; Felsenfeld, G. Conserved CTCF Insulator Elements Flank the Mouse and Human β-Globin Loci. Mol. Cell. Biol. 2002, 22, 3820–3831.
- Heger, P.; Marin, B.; Bartkuhn, M.; Schierenberg, E.; Wiehe, T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 17507–17512.
- Nora, E.P.; Caccianini, L.; Fudenberg, G.; So, K.; Kameswaran, V.; Nagle, A.; Uebersohn, A.; Hajj, B.; Le Saux, A.; Coulon, A.; et al. Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 2020, 11, 1–13.
- Parelho, V.; Hadjur, S.; Spivakov, M.; Leleu, M.; Sauer, S.; Gregson, H.C.; Jarmuz, A.; Canzonetta, C.; Webster, Z.; Nesterova, T.; et al. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell 2008, 132, 422–433.
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049.
- Garcia-Luis, J.; Lazar-Stefanita, L.; Gutierrez-Escribano, P.; Thierry, A.; Cournac, A.; García, A.; González, S.; Sánchez, M.; Jarmuz, A.; Montoya, A.; et al. FACT mediates cohesin function on chromatin. Nat. Struct. Mol. Biol. 2019, 26, 970–979.




