
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ana Banito | + 1610 word(s) | 1610 | 2021-04-12 05:01:51 | | | |
| 2 | Vivi Li | Meta information modification | 1610 | 2021-04-13 04:38:20 | | |
Video Upload Options
Sarcomas are mesenchymal malignancies accounting for about 15% of cancers in children and adolescents, making them the third most common group of childhood cancers, following blood malignancies and brain tumors.
1. Introduction
Sarcomas are mesenchymal malignancies accounting for about 15% of cancers in children and adolescents, making them the third most common group of childhood cancers, following blood malignancies and brain tumors [1]. While the last decades have seen vast improvements in pediatric cancer care with overall improved prognosis, this does not hold true for sarcomas, which are often prone to metastasis and relapse, typically accompanied by dismal prognosis [2]. Research efforts to improve this situation are complicated by the extremely diverse intrinsic nature of pediatric sarcoma with more than 60 genetically distinct entities [3]. While some pediatric sarcoma types may show widespread genomic instability (e.g., osteosarcoma (OS)), many are genetically rather simple, characterized by pathognomonic fusion oncogenes (e.g., SSX-SS18 in Synovial Sarcoma (SySa)) [4][5][6]. Ongoing molecular profiling efforts will likely lead to further sub-classification as we learn more about specific genetic and epigenetic alterations and underlying biology [7]. Figure 1 provides a snapshot of the most common pediatric sarcoma types according to the recently (2020) updated World Health Organization (WHO) classification of soft tissue and bone tumors (Figure 1a) [8][9], as well as an unbiased molecular clustering of the most common tumor entities based on DNA methylation data (Figure 1b) [10]. The development of novel therapeutic agents heavily relies on preclinical testing in disease specific models. Given the rarity of each individual pediatric sarcoma subtype, appropriate model systems are naturally scarce, and the selection of suitable models is challenging. This represents a major problem for pediatric cancer research and has significantly contributed to the lack of meaningful therapeutic improvements in pediatric sarcomas [11]. Therefore, in this review, we aim to outline the pros and cons of major in vivo modeling approaches applicable for pediatric sarcoma. We review existing models applied for specific sarcoma entities and discuss relevant points to consider for meaningful future model utilization. Due to the vast array of known malignancies, the scope of this review is limited to 18 clinically particularly relevant sarcoma entities.
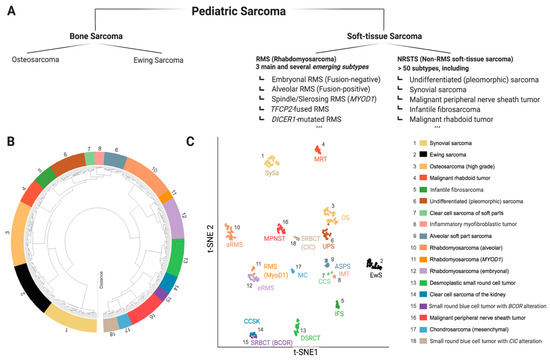
Figure 1. The diverse landscape of pediatric sarcoma. (A) Extraction from the current WHO classification of soft-tissue and bone tumors (5th edition, 2020) with focus on pediatric sarcoma [8]. (B,C) Molecular classification by whole genome DNA methylation data, here depicted for 18 sarcoma entities relevant for childhood and adolescence. Data shown as hierarchical clustering analysis, (B) and t-distributed stochastic neighbor embedding (t-SNE), (C) data adapted from Kölsche et al., 2021 [10].
2. In Vivo Modeling Approaches Applicable for Pediatric Sarcoma
Analogous to other solid tumors, four main general approaches of in vivo cancer modeling can be distinguished for pediatric sarcoma (Figure 2) [12][13]. Two of these entail the engraftment of human cancer tissue into immunocompromised mice—cell-line-derived xenograft models (CDXs) and patient-derived xenograft models (PDXs)—while the other two induce de novo tumorigenesis in immunocompetent wild type mice—environmentally-induced models (EIMMs) and genetically-engineered mouse models (GEMMs). Figure 3 highlights the major advantages and disadvantages of these approaches. Independent of the specific approach, one should consider that different mouse strains, much like humans, possess an inherent and strain-specific risk of spontaneously developing different cancer entities over their lifespans [14][15]. While these can, in some cases, also serve as useful models of human cancer, they should by no means be mistaken for specifically engrafted human or induced murine tumor tissue [16].
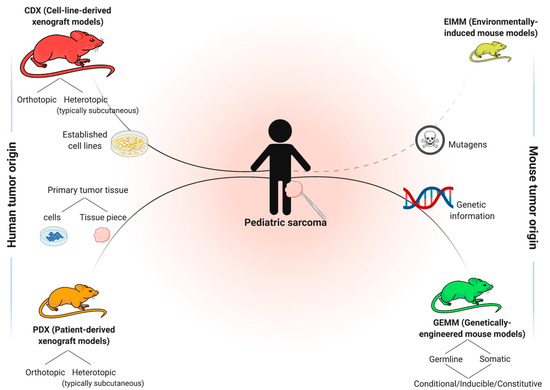
Figure 2. Different approaches to model sarcoma in vivo. Relative sizes of mouse pictograms resemble approximate utilization of modeling approaches in current sarcoma research. Dashed line illustrates that environmentally-induced models (EIMMs) are typically not particularly relevant for childhood sarcoma.
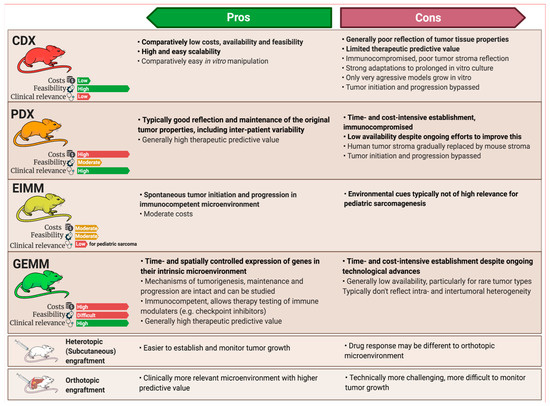
Figure 3. Pros and cons of different in vivo modeling approaches. While cell-line-derived xenograft models (CDXs) and patient-derived xenograft models (PDXs) are both engraftment models, environmentally-induced models (EIMMs) and genetically-engineered mouse models (GEMMs) can be utilized to establish syngeneic engraftment models (SAMs), enabling scalability for these models, too. Pros and cons of different engraftment sites depicted at the bottom apply to all of the engraftment models, regardless of origin.
CDXs are the most commonly used, but least representative model when aiming to recapitulate the original disease. EIMMs are extremely powerful, but since pediatric sarcomas are usually not driven by environmental factors, they are not as relevant for childhood sarcoma. PDXs and GEMMs however, are highly representative of their human counterpart, therapeutically predictive and complementary to each other in nature.
3. Applications of Pediatric Sarcoma Mouse Models
Most and foremost, model generation is no end in itself. Both biological and translational advances require purposeful utilization of the right model system for the respective research question. While CDXs are still the by far most commonly used model due to broad availability and ease of use, either PDX- or GEM-models are typically the most suitable model for both basic and translational research questions (Figure 4). In general, GEM models are ideal to deepen our understanding of basic sarcoma biology while PDX models are of particularly value for preclinical testing, adequately representing patient heterogeneity. While many cell-based immunotherapies can also be tested in conventional PDX models, immunotherapies requiring endogenous immune cells can either be tested in GEMMs or humanized PDX models, the latter being very costly and technically challenging thus largely limiting their use [17][18] (Figure 5b). Both, PDX and GEM models are suitable for local therapy advancement and imaging studies, a rather underrepresented field of research, given the importance of complete resection for clinical outcome [19][20].
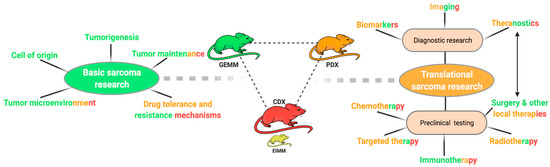
Figure 4. Utilization of sarcoma mouse models. Schematic overview of different research questions in sarcoma research. Color codes represent the generally most suitable modeling approach for the respective research field ((green) = GEMM, (orange) = PDX, (red) = CDX).
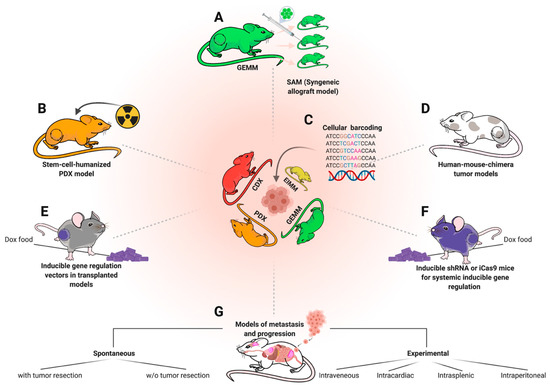
Figure 5. Complementary in vivo modeling approaches. The depicted methods are not specific to sarcoma, but can be applied to complement and optimize utilization of existing models and approaches. (A) Derivation of syngeneic engraftment models (SAMs) from GEMMs to increase scalability. (B) Humanization of existing PDX models for immunotherapy trials requiring an endogenous immune system, e.g., checkpoint inhibitor therapies. (C) Cellular barcoding of engraftment models to study clonal selection effects under therapy and metastasis. (D) Human mouse chimera genetic models to bypass mouse-human biology discrepancies. (E) Transduction of tumor cells from engraftment models using Doxycycline (Dox)-inducible vectors (e.g., TRE-shRNA to inducible knockdown a gene of interest) to investigate molecular dependencies in vivo. (F) Applying the same approach (E) on a systemic level to additionally study systemic toxicity in a target-gene-dependent fashion. (G) Overview of divergent metastasis modeling approaches.
GEMMs of different genetic makeup can also be used to assess the fraction of tumor cells with tumorigenic potential, following the notion that some tumors rely on a small fraction of cells to drive overall cell renewal and tumor growth [21]. Following this cancer stem cell idea, Buchstaller et al., for example, compared the tumorigenic potential of two engrafted MPNST GEMMs and found that transplanted MPNST cells from Nf1+/− Ink4a/Arf−/− tumors encompassed a 10-fold higher fraction of cells with cancer-initiating potential than MPNST cells from Nf1+/− and Tp53+/− tumors [22].
Given these advantages of PDXs and GEMMs, CDXs possess one natural prime advantage GEMMs and PDXs are often lacking: they entail a corresponding in vitro system, allowing for variable functional characterization and are often very well characterized. This strength paired with the high practicality of their use makes them a highly valuable tool for present and future sarcoma research.
Considerations for Preclinical Testing
A major consideration for model utilization is how to design meaningful and translatable preclinical therapy trials. This is particularly important for pediatric sarcoma since the rarity of individual subtypes combined with the incredibly diverse array of subsets complicates rational clinical trial design. To this end, Langenau et al. provided a comprehensive and contemporary review, highlighting 10 key points to consider when designing preclinical treatment trials [23]. The key concept is to apply the same principles and guidelines used in clinical phase I, II, and III trials to the preclinical setting in a similar stepwise approach by conducting preclinical phase I, II and III trials alike [23]. This systematic approach is equally feasible for combinatorial agent testing in vivo and elucidated that some synergistic effects can be mediated by the in vivo environment and are not picked up in vitro [24]. A prerequisite of sublime importance for successful in vivo trials is the appropriate selection of promising treatment agents, based on comprehensive molecular and drug-screening in vitro data [25]. Equally important is the selection of a set of appropriate and well characterized model systems, adequately representing patient heterogeneity, including relevant patient subsets based on molecular characteristics serving as biomarkers, possibly informing about treatment response [26][27]. Connecting molecular model characterization and drug response data is an essential avenue in moving towards precision oncology in pediatrics [26]. Recent reports applying this concept by conducting single-mouse-design studies highlight the feasibility and translational relevance of this approach [28][29][30]. Approaches to use PDX models as avatars for individual patients that are parallelly being treated in the clinic are possible in principle, but typically hampered by time-consuming model establishment and variable engraftment rates [31]. Nimmervoll et al. further introduced the concept of a mouse clinic, aiming to more closely resemble the multimodal clinical therapy, including chemotherapy, radiotherapy, and local resection for testing the application of new targeted treatments [32]. While this elaborate approach will likely be to too complex and resource-intensive for general use, one should carefully consider the combination of new targeted treatments and immunotherapies with mainstays of current therapy, including local resection and radiation. A more feasible approach to deepen the insight of therapy trials is the use of molecular barcoding of engrafted cells to reveal therapy-induced clonal selection processes [33][34][35] (Figure 5c). Useful examples on how to present preclinical therapy response data can be found in the review from Gengenbacher et al. [13].
References
- Allen-Rhoades, W.; Whittle, S.B.; Rainusso, N. Pediatric Solid Tumors in Children and Adolescents: An Overview. Pediatr. Rev. 2018, 39, 444–453.
- Sandler, G.; Yokoi, A.; Hayes-Jordan, A. An update in the management of pediatric sarcoma. Curr. Opin. Pediatr. 2019, 31, 368–377.
- Cao, J.; An, Q.; Wang, L. Pediatric sarcomas. Oncol. Lett. 2018, 15, 1397–1402.
- Nakano, K.; Takahashi, S. Translocation-Related Sarcomas. Int. J. Mol. Sci. 2018, 19, 3784.
- Miettinen, M.; Felisiak-Golabek, A.; Luiña Contreras, A.; Glod, J.; Kaplan, R.N.; Killian, J.K.; Lasota, J. New fusion sarcomas: Histopathology and clinical significance of selected entities. Hum. Pathol. 2019, 86, 57–65.
- Laetsch, T.W.; Roy, A.; Xu, L.; Black, J.O.; Coffin, C.M.; Chi, Y.Y.; Tian, J.; Spunt, S.L.; Hawkins, D.S.; Bridge, J.A.; et al. Undifferentiated sarcomas in children harbor clinically relevant oncogenic fusions and gene copy-number alterations: A report from the children’s oncology group. Clin. Cancer Res. 2018, 24, 3888–3897.
- Shern, J.F.; Chen, L.; Chmielecki, J.; Wei, J.S.; Patidar, R.; Rosenberg, M.; Ambrogio, L.; Auclair, D.; Wang, J.; Song, Y.K.; et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014, 4, 216–231.
- WHO Classification of Tumours Editorial Board (Ed.) World Health Organization Classification of Soft Tissue and Bone Tumours, 5th ed.; IARC Press: Lyon, France, 2020.
- Anderson, W.J.; Doyle, L.A. Updates from the 2020 World Health Organization Classification of Soft Tissue and Bone Tumours. Histopathology 2021, his.14265.
- Koelsche, C.; Schrimpf, D.; Stichel, D.; Sill, M.; Sahm, F.; Reuss, D.E.; Blattner, M.; Worst, B.; Heilig, C.E.; Beck, K.; et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 2021, 12, 498.
- Jones, D.T.W.; Banito, A.; Grünewald, T.G.P.; Haber, M.; Jäger, N.; Kool, M.; Milde, T.; Molenaar, J.J.; Nabbi, A.; Pugh, T.J.; et al. Molecular characteristics and therapeutic vulnerabilities across paediatric solid tumours. Nat. Rev. Cancer 2019, 19, 420–438.
- Day, C.-P.; Merlino, G.; Van Dyke, T. Preclinical mouse cancer models: A maze of opportunities and challenges. Cell 2015, 163, 39–53.
- Gengenbacher, N.; Singhal, M.; Augustin, H.G. Preclinical mouse solid tumour models: Status quo, challenges and perspectives. Nat. Rev. Cancer 2017, 17, 751–765.
- Hunter, K.W. Mouse models of cancer: Does the strain matter? Nat. Rev. Cancer 2012, 12, 144–149.
- Puccini, J.; Dorstyn, L.; Kumar, S. Genetic background and tumour susceptibility in mouse models. Cell Death Differ. 2013, 20, 964.
- Moyer, A.M.; Yu, J.; Sinnwell, J.P.; Dockter, T.J.; Suman, V.J.; Weinshilboum, R.M.; Boughey, J.C.; Goetz, M.P.; Visscher, D.W.; Wang, L. Spontaneous murine tumors in the development of patient-derived xenografts: A potential pitfall. Oncotarget 2019, 10, 3924–3930.
- Sanmamed, M.F.; Chester, C.; Melero, I.; Kohrt, H. Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 1190–1198.
- Tian, H.; Lyu, Y.; Yang, Y.G.; Hu, Z. Humanized Rodent Models for Cancer Research. Front. Oncol. 2020, 10.
- Buchakjian, M.R.; Merritt, N.M.; Moose, D.L.; Dupuy, A.J.; Tanas, M.R.; Henry, M.D. A Trp53fl/flPtenfl/fl mouse model of undifferentiated pleomorphic sarcoma mediated by adeno-Cre injection and in vivo bioluminescence imaging. PLoS ONE 2017, 12, e0183469.
- Chen, C.; Dorado Garcia, H.; Scheer, M.; Henssen, A.G. Current and Future Treatment Strategies for Rhabdomyosarcoma. Front. Oncol. 2019, 9, 1–18.
- Atashzar, M.R.; Baharlou, R.; Karami, J.; Abdollahi, H.; Rezaei, R.; Pourramezan, F.; Zoljalali Moghaddam, S.H. Cancer stem cells: A review from origin to therapeutic implications. J. Cell. Physiol. 2020, 235, 790–803.
- Joseph, N.M.; Mosher, J.T.; Buchstaller, J.; Snider, P.; McKeever, P.E.; Lim, M.; Conway, S.J.; Parada, L.F.; Zhu, Y.; Morrison, S.J. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 2008, 13, 129–140.
- Langenau, D.M.; Sweet-Cordero, A.; Wechsler-Reya, R.; Dyer, M.A. Preclinical Models Provide Scientific Justification and Translational Relevance for Moving Novel Therapeutics into Clinical Trials for Pediatric Cancer. Cancer Res. 2015, 75, 5176–5186.
- Houghton, P.J.; Morton, C.L.; Gorlick, R.; Lock, R.B.; Carol, H.; Reynolds, C.P.; Kang, M.H.; Maris, J.M.; Keir, S.T.; Kolb, E.A.; et al. Stage 2 combination testing of rapamycin with cytotoxic agents by the pediatric preclinical testing program. Mol. Cancer Ther. 2010, 9, 101–112.
- Stewart, E.; Federico, S.; Karlstrom, A.; Shelat, A.; Sablauer, A.; Pappo, A.; Dyer, M.A. The Childhood Solid Tumor Network: A new resource for the developmental biology and oncology research communities. Dev. Biol. 2016, 411, 287–293.
- DuBois, S.G.; Corson, L.B.; Stegmaier, K.; Janeway, K.A. Ushering in the next generation of precision trials for pediatric cancer. Science 2019, 363, 1175–1181.
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95.
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325.
- Murphy, B.; Yin, H.; Maris, J.M.; Kolb, E.A.; Gorlick, R.; Reynolds, C.P.; Kang, M.H.; Keir, S.T.; Kurmasheva, R.T.; Dvorchik, I.; et al. Evaluation of alternative in vivo drug screening methodology: A single mouse analysis. Cancer Res. 2016, 76, 5798–5809.
- Ghilu, S.; Li, Q.; Fontaine, S.D.; Santi, D.V.; Kurmasheva, R.T.; Zheng, S.; Houghton, P.J. Prospective use of the single-mouse experimental design for the evaluation of PLX038A. Cancer Chemother. Pharmacol. 2020, 85, 251–263.
- Stebbing, J.; Paz, K.; Schwartz, G.K.; Wexler, L.H.; Maki, R.; Pollock, R.E.; Morris, R.; Cohen, R.; Shankar, A.; Blackman, G.; et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 2014, 120, 2006–2015.
- Nimmervoll, B.V.; Boulos, N.; Bianski, B.; Dapper, J.; DeCuypere, M.; Shelat, A.; Terranova, S.; Terhune, H.E.; Gajjar, A.; Patel, Y.T.; et al. Establishing a Preclinical Multidisciplinary Board for Brain Tumors. Clin. Cancer Res. 2018, 24, 1654–1666.
- Merino, D.; Weber, T.S.; Serrano, A.; Vaillant, F.; Liu, K.; Pal, B.; Di Stefano, L.; Schreuder, J.; Lin, D.; Chen, Y.; et al. Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer. Nat. Commun. 2019, 10.
- Seth, S.; Li, C.Y.; Ho, I.L.; Corti, D.; Loponte, S.; Sapio, L.; Del Poggetto, E.; Yen, E.Y.; Robinson, F.S.; Peoples, M.; et al. Pre-existing Functional Heterogeneity of Tumorigenic Compartment as the Origin of Chemoresistance in Pancreatic Tumors. Cell Rep. 2019, 26, 1518–1532.e9.
- Echeverria, G.V.; Powell, E.; Seth, S.; Ge, Z.; Carugo, A.; Bristow, C.; Peoples, M.; Robinson, F.; Qiu, H.; Shao, J.; et al. High-resolution clonal mapping of multi-organ metastasis in triple negative breast cancer. Nat. Commun. 2018, 9.




