
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kyle Lauersen | + 2443 word(s) | 2443 | 2021-04-09 07:52:10 | | | |
| 2 | Dean Liu | -38 word(s) | 2405 | 2021-04-12 09:39:55 | | |
Video Upload Options
Microalgae and cyanobacteria are photosynthetic microbes that can be grown with the simple inputs of water, carbon dioxide, (sun)light, and trace elements.
1. Introduction
Microalgae and cyanobacteria are interesting study organisms capable of photosynthetic growth on carbon dioxide (CO2) as a sole carbon source. These organisms naturally synthesize different metabolites such as pigments, oils and lipids, sterols, starches, polysaccharides, and halogenated compounds, and algae are also amenable to contained, scalable growth in photobioreactors driven by (sun)light energy [1][2]. Genetic engineering of algae and cyanobacteria in the era of synthetic biology holds the promise of tailored production of novel and customized metabolites using sustainable waste CO2 as a feedstock. However, algae are a diverse and polyphyletic group of organisms which do not share close evolutionary relatedness and exhibit incredible variability in their genomes [3]. These features have hindered intensive molecular tool development except in a handful of model species, and their broader application to biotechnology has been slower compared to other hosts such as bacteria, yeast, plant, and mammalian cells [2][4].
The genomic diversity of algae necessitates customized molecular tools that work with genetic architecture of a specific host. Once these tools are produced by piecemeal cloning or complete DNA synthesis, the reliable introduction of foreign genetic material into cells is an essential prerequisite to biotechnological concepts [5][6][7]. DNA delivery methods result in stable chromosomal integration or episomal/plasmid extrachromosomal replication of foreign transgene expression elements [7][8][9]. Methodologies of DNA delivery and cell membrane/wall permeabilization vary according to the host organism and target cellular compartment (organellar or nuclear). For example, some cyanobacteria will natively take up DNA from their environment without the need for manipulation, while many eukaryotic algae maintain cell walls that necessitate more aggressive delivery methods [10][11].
Foreign DNA delivery can be achieved by mechanical agitation, surfactant permeabilization, electroporation, particle bombardment, and bacterial DNA transfer (conjugation or Agrobacterium tumefaciens-mediated) [9][12][13][14][15]. In addition to these classical methods, emerging techniques used in other cell systems that have not been widely applied to algae may potentially improve DNA delivery (Figure 1). Strategies based on cell penetrating peptides (CPP), polymers, metal-organic frameworks, nanoparticles, and liposomes have all been demonstrated in non-algal hosts [16][17][18]. This review examines recent progress in gene delivery methodologies and discusses their technical aspects, advantages, limitations, and potential in the context of algal biotechnology.
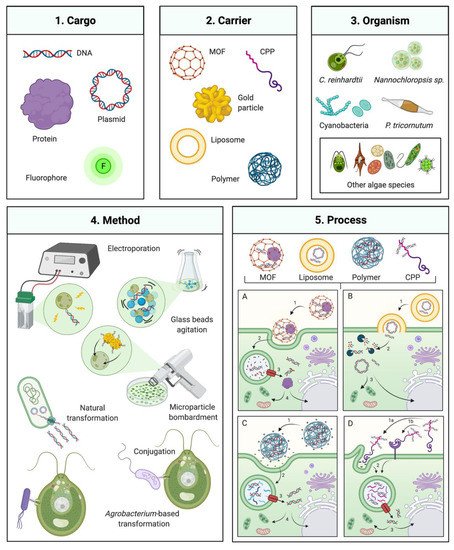
Figure 1. Gene delivery technologies for microalgae engineering. Overview of several available transformation technologies which have been or could be applied to algal engineering. (1) and (2) Several carriers that can mediate transformation of a DNA, protein, or chemical cargo (left panel) MOF: metal-organic framework, CPP: Cell-penetrating peptide. (3) Algal species which transformation has been commonly employed are depicted on the upper right: C. reinhardtii, Nannochloropsis sp., and P. tricornutum, including various cyanobacteria and other algal species as described in the manuscript. (4) Classical transformation methods are presented in the bottom left panel. (5) Process of transformation mediated by different DNA carriers and their interaction with the cell for delivery of the respective cargo. (5A) MOF-DNA, (5A1) recognition and cell uptake by endocytosis, (5A2) internalization and fusion with phagolysosome, 3 phagolysosome escape, 4 transport to nucleus, chloroplast or mitochondria; (5B) Liposome-DNA, (5B1) lyposome integration with cell membrane, (5B2) cargo (DNA or protein) exposed in cytoplasm and potential enzymatic degradation, 3 intact cargo transport to nucleus, mitochondria or chloroplast; (5C) Polymer-DNA, (5C1) cell recognition of polymer nanoparticle, charge interaction and cell uptake by endocytosis, (5C2) internalization and fusion with phagolysosome, (5C3) phagolysosome escape, (5C4) transport to nucleus, chloroplast or mitochondria; (5D) CPP-DNA, 1 cell recognition of CPP, by charge interaction (5D1a) or receptor recognition (5D1b) and cell uptake by endocytosis, (5D2) internalization and fusion with phagolysosome, (5D3) phagolysosome escape, (5D4) Transport to nucleus, chloroplast or mitochondria. Created with BioRender.com.
2. Traditional Algal Transformation Techniques
2.1. Agitation of Cells in the Presence of DNA and Non-Ionic Surfactants
The agitation of algal cells in the presence of glass beads, polyethylene glycol (PEG), and foreign DNA has become a standard protocol for the delivery of foreign DNA into the model green microalgae C. reinhardtii and other species as Cyanidioschyzon merolae, Chlorella vulgaris, and Dunaliella salina [14][19][20][21]. This protocol has been adopted for nuclear and chloroplast transformation and can be applied to cell wall deficient cell lines or those with a cell wall when chemo/enzymatic removal is performed before transformation (as with Chlorella strains) [13][14][15][22][23][24]. Agitation-based transformation is achieved by agitation (vortexing) of cells in the presence of a physical agitator (glass beads), a non-ionic surfactant (i.e., polyethylene glycol (PEG)), and DNA [14][25]. Transformation efficiency depends on many factors such as cell size, presence or absence of a cell wall, duration of agitation, velocity, the concentration of surfactant, and the use of linear or circular DNA [26]. The frequency of DNA integration into the C. reinhardtii genome with glass bead agitation has been reported at ~103 transformants μg of DNA-1 with ~108 cells mL−1 starting cell concentration [14]. Reports have shown that numerous other non-reactive materials, such as silicon carbide whiskers can also serve this purpose [27]. Agitation-based transformation protocols are advantageous as they do not require specialized equipment, and are inexpensive and relatively fast [14][15].
A significant limitation of the technique is the requirement of cell wall removal, which can be achieved with the autolysin protein of C. reinhardtii for itself, or other enzyme cocktails (e.g., cellulase, macerase, pectinase, and hemicellulose) for other hosts [14][19][28][29][30][31][32]. Reports have shown that cell-wall free protoplasts of Chlorella ellipsoidea and the naturally cell wall deficient C. merolae have been transformed using DNA and PEG without mechanical agitation, suggesting that surfactant mediated permeabilization is sufficient to enable DNA uptake [33]. However, the main drawback of agitation-based transformation techniques is the low transformation efficiency [34]. This has led to development of other more efficient strategies.
2.2. Electroporation
DNA transformation by electroporation is as an alternative method in numerous algae and pioneered in C. reinhardtii [34]. This technique uses electrodes to generate voltage differential across the cell membrane, temporarily disturbing the phospholipid bilayer, allowing molecules to pass into the cell [35][36]. Transformation efficiencies from electroporation vary according to factors such as field-strength, pulse length, ionic strength of medium composition, temperature, cell membrane characteristics, the species used, its molecular toolkit, and the presence of cell-wall [36][37]. Electroporation can increase transformation efficiency up to 100-fold over agitation and is not affected by the cell wall. Fewer sequence deletions have been observed at the genomic insertion sites than those seen from agitation transformation [34][37][38]. Electroporation has been shown to enable the transformation of cell-wall containing algae and cyanobacteria such as Monoraphidium neglectum, Nannochloropsis sp., Phaeodactylum tricornutum, C. reinhardtii, Anabaena, and Nostoc punctiforme [34][35][39][40][41][42][43][44]. Electroporation is a useful technique for delivering DNA, RNA, proteins, nucleotides, and dyes into cells [34][35][45]. Electroporation is a rapid protocol and requires equipment which is often present in microbial laboratories to transform bacterial cells [37][46].
Electroporation efficiencies were increased by considering the specific mitotic phase of C. reinhardtii growth [47]. Synchronization of C. reinhardtii cultures has been shown to increase transformation efficiency after three days. C. reinhardtii culture synchronization using 28 °C during the light phase and 18 °C in the dark phase was also shown to enable increased homologous recombination efficiencies specifically at 12 hours after illumination initiation [48]. Digital microfluidics (DME) have also been used to optimize electroporation protocols, using a fluid mixture of cell/DNA droplets encapsulated in biocompatible oil, and electric pulses applied from an array of microelectrodes to the droplets [49][50]. DME showed an efficiency of 2.5 × 104 of C. reinhardtii mutants per μg of DNA without cell wall removal with an initial concentration of 2 × 106 cells mL−1 algal cells, and 1 μg DNA [49]. A square-electric-pulse electroporation technique has also been reported with efficiencies of 6 × 103 C. reinhardtii transformants per μg of DNA with 1.5 × 107 cells mL−1 initial cell density 0.1 μg of DNA [47]. However, this technique has not been broadly adopted, as it requires specialized equipment. It was described in N. limnetica, which contains a rigid cell wall, that treatment with 10 mM lithium acetate and 3 mM dithiothreitol (DTT) before electroporation improved transformation efficiency by increasing cell wall permeability, resulting in 1.1 × 107 transformants per μg of DNA with an initial concentration of 3.3 × 109 cells mL−1 and 4 μg of DNA [40].
2.3. Microparticle Bombardment
Microparticle bombardment is one of the most versatile gene delivery methods due to its ability to transform the nucleus, mitochondria, or chloroplast genomes and even transform cells containing cell walls [13][51][52][53]. Microparticle bombardment is based on accelerated non-reactive metal (gold or tungsten) micro-projectiles coated with DNA being shot at and colliding with target cells [1][51]. The impact of these particles allows them to penetrate the cells and deliver foreign DNA. Transformation efficiency depends on the starting cell density, target organelle, selection efficiency, the number of DNA-coated particles, the DNA concentration on each particle, the kinetic energy of the particles, temperature, and ability of the cell to regenerate after particle damage [54].
The most common use for this technique is the transformation of chloroplast genomes, as it enables penetration of DNA through multiple membrane layers, but it has also recently been used to transform circular plasmids into the nucleus of the red alga Porphyridium purpureum [23][51][55]. Unlike agitation, but similar to electroporation, cell-wall presence does not affect transformation efficiency; however, cell viability can be disturbed if many micro-projectiles are used [51][56]. The efficiency of nuclear transformation in C. reinhardtii has been as low as 15 transformants per μg of DNA, with ~107 cells mL−1 starting cell concentration and 0.8 μg of DNA [13]. Efficiencies reported in D. salina, Volvox carteri and P. tricornutum are 2.5 × 10−5, 1.7 × 102, and 1.0 × 102 transformants per μg of DNA (0.1–0.7 μg of DNA), respectively, with 105–107 cells mL−1 as starting cell density [57][58][59]. However, it was reported that in C. reinhardtii, transformation efficiency could be improved when smaller particles are used (0.6 µm vs. 1.0 µm) [60].
Microparticle bombardment has also been described for the delivery of 24–68 kDa proteins, a technique called proteolistics. This approach is a simple physical deposition of target protein onto delivery substrate which is then used as other microprojectiles [61]. The application was shown to deliver Cas9–gRNA ribonucleoprotein (RNP) into P. tricornutum, where knock out of the PtUMPS and PtAPT genes led to 5-fluoroorotic acid (5-FOA) and 2-fluoroadenine (2-FA) resistance at an efficiency of ~10−6 per 106 cells mL−1 [62]. This technique was also found to be suitable for multiplexing mutations [62] and could become a powerful strategy for generating targeted knockouts in this and other diatoms/algae.
One disadvantage of bombardment is the requirement of gene-gun infrastructure as well as reagents and equipment. Transformation parameters must also be optimized for every alga and target cell compartment/genome [9][63]. One thing that must be considered is the potential post-transcriptional gene silencing (PTGS) due to the relatively high copy number of transgenes integrated into the genome [53][64].
3.2. Bacterial Conjugation
Conjugation is based on the ability of bacteria to share genetic information by exchanging plasmids through pili. This natural process can be employed using either a double or triple vector approach combining shuttle, conjugative, and helper plasmids. The shuttle and helper plasmids mediate the transfer of a conjugative plasmid between E. coli and the target host while the helper plasmid aids transformation by preventing vector degradation by endogenous restriction systems; it codes for DNA methylases (Aval, AvaW, and AvaIII) that prevent target host restriction enzyme recognition [65][66]. The helper vector contains an oriT-region, bom-site, and mob genes that encode a nickase and enable conjugation [67][68]. Bacterial conjugation has been used commonly in cyanobacterial species such as Synechococcus, Prochlorococcus, N. punctiforme, Anabaena, and Synechocystis sp. and recently in eukaryotic algae such as P. tricornutum, Thalassiosira pseudonana, Acutodesmus obliquus, and Neochloris oleoabundans [66][68][69][70][71][72][73][74][75]. The effectiveness of transformation by conjugation is based on the capability of the algal recipient strain to integrate and maintain the vector, either in the chromosome, in endogenous plasmids, or as an episomal plasmid [65][66][74][75].
Episomes, circular plasmids which self-replicate and do not integrate into the chromosome, can be efficient and straightforward vectors to enable transgene delivery of desired genetic elements between bacteria, cyanobacteria, and eukaryotes. Episomes avoid insertions and knock-out of non-targets and replicate independently from chromosomes [76][77][78]. Yeast centromere sequences (CEN/ARS) were found to act as autosomal replicating elements that allow stable maintenance of the circular episomal plasmid in diatoms [66][79]. Factors such as the growth phase of both target and donor organisms are crucial factors for proper conjugation [80]. Episomal vectors to transform microalgae via conjugation provide an efficient means of multi-gene pathway transfer due to stable self-replication of episomal vector and minimal possibility of positional or epigenetic effects [66]. Conjugation has the advantage over electroporation, microparticle bombardment, and glass-beads agitation to deliver larger DNA fragments [45]. The use of the conjugation-based method in P. tricornutum, N. oceanica, and Neochloris oleoabundans has been demonstrated to generate mutants with higher transformation efficiency compared to microparticle bombardment transformation: 500–1000 transformants/108 cells/200 ng DNA (conjugation) compared to 5–25 transformants/108 cells/2.5 µg DNA (bombardment) in P. tricornutum [74][81][82][83].
3.3. Agrobacterium-Mediated Transformation
The use of A. tumefaciens to transform plant cells relies on the natural infection process of the bacterium, which causes crown gall tumors on various plant species [84]. Natural tumor formation is a consequence of replicating a single-stranded copy of the transferred bacterial tumor-inducing (Ti) plasmid. The Ti plasmid and A. tumefaciens have been exploited since 1988 when it was found to allow transfer and permit stable integration of DNA fragments into a target higher plant genomes and has even been shown to transform mammalian cells [85][86][87]. The technique has the reported advantage of low gene rearrangements and foreign transcript silencing in plant cell lines [88][89].
A. tumefaciens-based transformation of algae has been reported in several species, although its use has not been wildly adopted. Reports exist of transformation by Agrobacterium method in C. reinhardtii, H. lacustris, Chlorella sp., Dunaliella bardawil, Symbiodinium sp., Nannochloropsis sp. and Parachlorella kessleri [85][90][91][92][93][94]. However, questions remain about how the bacterium, which evolved to infect plant cells, can infect evolutionarily distant algae and whether the infection requires specific recognition machinery on the target, or whether it is target independent. The standard protocol for Agrobacterium-mediated transformation is to mix target cell cultures with Agrobacterium containing a transgene cassette of interest; the mixture is exposed with the virulence agent acetosyringone, which signals the bacterium to infect wounded plant tissue. After transformation of the target cell by the bacterium, the selection is made using an antibiotic that can be selective against the bacterium while selecting the transformed cells. Variable transformation efficiencies have been reported depending on the protocol followed [90][95][96][97]. Factors such as temperature, pH, and time of virulence gene induction have been reported to have a substantial effect on transformation efficiency [98]. This method may enable integrating multigene pathways into host genomes as the capacity of the Ti plasmid has been reported as large as 150 kbp. If stably integrated into the genome, this would vastly outperform current methods of gene delivery that exhibit random nuclease digest en route to the nucleus, especially in green algae [99][100][101]. However, a recent report indicated that this method is no better than electroporation for stable integration of a plasmid containing a luciferase expression cassette and a selectable marker cassette into Parachlorella kessleri [90]. Further investigation is required to determine if this standard plant transformation protocol can be robustly applied to enhancing multi-gene expression cassette delivery to algal hosts.
References
- Vioque, A. Transformation of cyanobacteria. Adv. Exp. Med. Biol. 2007, 616, 12–22.
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 2015, 6, 1–10.
- Currier, T.C.; Wolk, C.P. Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J. Bacteriol. 1979.
- Thiel, T.; Peter Wolk, C. Conjugal Transfer of Plasmids to Cyanobacteria. Methods Enzymol. 1987, 153, 232–243.
- Wolk, C.P.; Vonshak, A.; Kehoe, P.; Elhai, J. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Isotopenpraxis 1984, 20, 1561–1565.
- Brahamsha, B. A Genetic Manipulation System for Oceanic Cyanobacteria of the Genus Synechococcus. Appl. Environ. Microbiol. 1996, 62, 1747–1751.
- Tolonen, A.C.; Liszt, G.B.; Hess, W.R. Genetic Manipulation of Prochlorococcus Strain MIT9313: Green Fluorescent Protein Expression from an RSF1010 Plasmid and Tn5 Transposition. Appl. Environ. Microbiol. 2006, 72, 7607–7613.
- Marraccini, P.; Bulteau, S.; Cassier-Chauvat, C.; Mermet-Bouvier, P.; Chauvat, F. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 1993, 23, 905–909.
- Tsinoremas, N.F.; Kutach, A.K.; Strayer, C.A.; Golden, S.S. Efficient gene transfer in Synechococcus sp. strains PCC 7942 and PCC 6301 by interspecies conjugation and chromosomal recombination. J. Bacteriol. 1994, 176, 6764–6768.
- Muñoz, C.F.; Sturme, M.H.J.; D’Adamo, S.; Weusthuis, R.A.; Wijffels, R.H. Stable transformation of the green algae Acutodesmus obliquus and Neochloris oleoabundans based on E. coli conjugation. Algal Res. 2019, 39, 101453.
- Fabris, M.; George, J.; Kuzhiumparambil, U.; Lawson, C.A.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Vickers, C.E.; Ralph, P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 2020.
- Dunahay, T.G.; Jarvis, E.E.; Roessler, P.G. Genetic Transformation of the Diatoms Cyclotella Cryptica and Navicula Saprophila. J. Phycol. 1995, 31, 1004–1012.
- Falciatore, A.; Casotti, R.; Leblanc, C.; Abrescia, C.; Bowler, C. Transformation of Nonselectable Reporter Genes in Marine Diatoms. Mar. Biotechnol. 1999, 1, 239–251.
- Miyagawa, A.; Okami, T.; Kira, N.; Yamaguchi, H.; Ohnishi, K.; Adachi, M. Research note: High efficiency transformation of the diatom Phaeodactylum tricornutum with a promoter from the diatom Cylindrotheca fusiformis. Phycol. Res. 2009, 57, 142–146.
- Diner, R.E.; Bielinski, V.A.; Dupont, C.L.; Allen, A.E.; Weyman, P.D. Refinement of the diatom episome maintenance sequence and improvement of conjugation-based DNA delivery methods. Front. Bioeng. Biotechnol. 2016, 4, 65.
- Cohen, M.F.; Wallis, J.G.; Campbell, E.L.; Meeks, J.C. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology 1994, 140, 3233–3240.
- Doron, L.; Segal, N.; Shapira, M. Transgene expression in microalgae—from tools to applications. Front. Plant Sci. 2016, 7, 505.
- Sharma, A.K.; Nymark, M.; Sparstad, T.; Bones, A.M.; Winge, P. Transgene-free genome editing in marine algae by bacterial conjugation—comparison with biolistic CRISPR/Cas9 transformation. Sci. Rep. 2018, 8, 1–11.
- Poliner, E.; Clark, E.; Cummings, C.; Benning, C.; Farre, E.M. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Res. 2020, 45, 101664.
- Poliner, E.; Takeuchi, T.; Du, Z.Y.; Benning, C.; Farre, E.M. Non-transgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779 Eric. Physiol. Behav. 2017, 176, 139–148.
- Tzfira, T.; Citovsky, V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr. Opin. Biotechnol. 2006, 17, 147–154.
- Kunik, T. Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA 2001, 98, 1871–1876.
- Schell, J.; Van Montagu, M. The Ti-plasmid of Agrobacterium tumefaciens, a natural vector for the introduction of nif genes in plants? Basic Life Sci. 1977, 9, 159–179.
- Zambryski, P.; Joos, H.; Genetello, C.; Leemans, J.; Van Montagu, M.; Schell, J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983, 2, 2143–2150.
- Hamilton, C.M.; Frary, A.; Lewis, C.; Tanksley, S.D. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. USA 1996, 93, 9975–9979.
- Valvekens, D.; Montagu, M.V.; Lijsebettens, M.V. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 1988, 85, 5536–5540.
- Kumar, S.V.; Misquitta, R.W.; Reddy, V.S.; Rao, B.J.; Rajam, M.V. Genetic transformation of the green alga - Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004, 166, 731–738.
- Kathiresan, S.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (chlorophyceae, volvocales). J. Phycol. 2009, 45, 642–649.
- Cha, T.S.; Yee, W.; Aziz, A. Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 2012, 28, 1771–1779.
- Rathod, J.P.; Prakash, G.; Pandit, R.; Lali, A.M. Agrobacterium-mediated transformation of promising oil-bearing marine algae Parachlorella kessleri. Photosynth. Res. 2013, 118, 141–146.
- Anila, N.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Establishment of Agrobacterium tumefaciens -mediated genetic transformation in Dunaliella bardawil. Eur. J. Phycol. 2011, 46, 36–44.
- Pratheesh, P.T.; Vineetha, M.; Kurup, G.M. An efficient protocol for the Agrobacterium-mediated genetic transformation of microalga Chlamydomonas reinhardtii. Mol. Biotechnol. 2014, 56, 507–515.
- Mini, P.; Demurtas, O.C.; Valentini, S.; Pallara, P.; Aprea, G.; Ferrante, P.; Giuliano, G. Agrobacterium-mediated and electroporation-mediated transformation of Chlamydomonas reinhardtii: A comparative study. BMC Biotechnol. 2018, 18, 11.
- Prasad, B.; Lein, W.; Thiyam, G.; Lindenberger, C.P.; Buchholz, R.; Vadakedath, N. Stable nuclear transformation of rhodophyte species Porphyridium purpureum: Advanced molecular tools and an optimized method. Photosynth. Res. 2019, 140, 173–188.
- Michielse, C.B.; Hooykaas, P.J.J.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17.
- Lauersen, K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta 2019, 249, 155–180.
- Zhang, R.; Patena, W.; Armbruster, U.; Gang, S.S.; Blum, S.R.; Jonikas, M.C. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 2014, 26, 1398–1409.
- Lin, H.; Cliften, P.F.; Dutcher, S.K. MAPINS, a highly efficient detection method that identifies insertional mutations and complex DNA rearrangements. Plant Physiol. 2018, 178, 1436–1447.
- Tang, D.K.H.; Qiao, S.Y.; Wu, M. Insertion mutagenesis of Chlamydomonas reinhardtii by electroporation and heterologous DNA. Biochem. Mol. Biol. Int. 1995, 36, 1025–1035.
- Chen, Y.; Hu, H. High efficiency transformation by electroporation of the freshwater alga Nannochloropsis limnetica. World J. Microbiol. Biotechnol. 2019, 35, 1–10.
- Zhang, C.; Hu, H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genom. 2014, 16, 63–66.
- Holmqvist, M.; Stensjö, K.; Oliveira, P.; Lindberg, P.; Lindblad, P. Characterization of the hupSL promoter activity in Nostoc punctiforme ATCC 29133. BMC Microbiol. 2009, 9.
- Thiel, T.; Poo, H. Transformation of a filamentous cyanobacterium by electroporation. J. Bacteriol. 1989, 171, 5743–5746.
- Jaeger, D.; Hübner, W.; Huser, T.; Mussgnug, J.H.; Kruse, O. Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum. J. Biotechnol. 2017, 249, 10–15.
- Doron, L.; Segal, N.; Shapira, M. Transgene expression in microalgae—from tools to applications. Front. Plant Sci. 2016, 7, 505.
- Yamano, T.; Iguchi, H.; Fukuzawa, H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 2013, 115, 691–694.
- Wang, L.; Yang, L.; Wen, X.; Chen, Z.; Liang, Q.; Li, J.; Wang, W. Rapid and high efficiency transformation of Chlamydomonas reinhardtii by square-wave electroporation. Biosci. Rep. 2019, 39.
- Angstenberger, M.; De Signori, F.; Vecchi, V.; Dall’Osto, L.; Bassi, R. Cell Synchronization Enhances Nuclear Transformation and Genome Editing via Cas9 Enabling Homologous Recombination in Chlamydomonas reinhardtii. ACS Synth. Biol. 2020, 9, 2840–2850.
- Im, D.J.; Jeong, S.-N.; Yoo, B.S.; Kim, B.; Kim, D.-P.; Jeong, W.-J.; Kang, I.S. Digital Microfluidic Approach for Efficient Electroporation with High Productivity: Transgene Expression of Microalgae without Cell Wall Removal. Anal. Chem. 2015, 87, 6592–6599.
- Bodénès, P.; Wang, H.-Y.; Lee, T.-H.; Chen, H.-Y.; Wang, C.-Y. Microfluidic techniques for enhancing biofuel and biorefinery industry based on microalgae. Biotechnol. Biofuels 2019, 12, 33.
- Boynton, J.E.; Gillham, N.W.; Harris, E.H.; Hosler, J.P.; Johnson, A.M.; Jones, A.R.; Randolph-Anderson, B.L.; Robertson, D.; Klein, T.M.; Shark, K.B.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538.
- Day, A.; Debuchy, R.; van Dillewijn, J.; Purton, S.; Rochaix, J.-D. Studies on the maintenance and expression of cloned DNA fragments in the nuclear genome of the green alga Chlamydomonas reinhardtii. Physiol. Plant. 1990, 78, 254–260.
- Remacle, C.; Cardol, P.; Coosemans, N.; Gaisne, M.; Bonnefoy, N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 2006, 103, 4771–4776.
- Altpeter, F.; Baisakh, N.; Beachy, R.; Bock, R.; Capell, T.; Christou, P.; Daniell, H.; Datta, K.; Datta, S.; Dix, P.J.; et al. Particle bombardment and the genetic enhancement of crops: Myths and realities. Mol. Breed. 2005, 15, 305–327.
- Li, Z.; Bock, R. Replication of bacterial plasmids in the nucleus of the red alga Porphyridium purpureum. Nat. Commun. 2018, 9, 1–8.
- Ortiz-Matamoros, M.F.; Villanueva, M.A.; Islas-Flores, T. Genetic transformation of cell-walled plant and algae cells: Delivering DNA through the cell wall. Brief. Funct. Genom. 2018, 17, 26–33.
- Schiedlmeier, B.; Schmitt, R.; Müller, W.; Kirk, M.M.; Gruber, H.; Mages, W.; Kirk, D.L. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 1994, 91, 5080–5084.
- Feng, S.; Xue, L.; Liu, H.; Lu, P. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 2009, 36, 1433–1439.
- Apt, K.E.; Grossman, A.R.; Kroth-Pancic, P.G. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. MGG 1996, 252, 572–579.
- Randolph-Anderson, B.; Boynton, J.E.; Dawson, J.; Dunder, E.; Eskes, R.; Gillham, N.W.; Johnson, A.; Perlman, P.S.; Suttie, J.; Heiser, W.C. Sub-Micron Gold Particles are Superior to Larger Particles for Efficient Biolistic Transformation of Organelles and Some Cell Types. 1995. Available online: (accessed on 31 January 2021).
- Martin-Ortigosa, S.; Wang, K. Proteolistics: A biolistic method for intracellular delivery of proteins. Transgenic Res. 2014, 23, 743–756.
- Serif, M.; Dubois, G.; Finoux, A.L.; Teste, M.A.; Jallet, D.; Daboussi, F. One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat. Commun. 2018, 9, 1–10.
- Mayfield, S.P.; Kindle, K.L. Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc. Natl. Acad. Sci. USA 1990, 87, 2087–2091.
- Ramesh, V.M.; Bingham, S.E.; Webber, A.N. A simple method for chloroplast transformation in Chlamydomonas reinhardtii. Methods Mol. Biol. 2011, 684, 313–320.




