
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Avri Ben-ze'ev | + 1389 word(s) | 1389 | 2021-04-08 08:19:51 | | | |
| 2 | Vivi Li | Meta information modification | 1389 | 2021-04-08 12:06:54 | | |
Video Upload Options
The Wnt/β-catenin cell–cell signaling pathway is one of the most basic and highly conserved pathways for intercellular communications regulating key steps during development, differentiation, and cancer. In colorectal cancer (CRC), in particular, aberrant activation of the Wnt/β-catenin pathway is believed to be responsible for perpetuating the disease from the very early stages of cancer development. A large number of downstream target genes of β-catenin-T-cell factor (TCF), including oncogenes, were detected as regulators of CRC development.
1. Introduction
Cell–cell adhesion is a basic biological process in multicellular organisms that determines cellular and tissue morphogenesis, and its disruption is a hallmark of cancer development. Aberrant signaling mediated by changes in cell–cell adhesion is a characteristic feature of invasive and metastatic cancer cells. Wnt/β-catenin signaling is a key signaling pathway that is hyperactivated in the majority of inherited colorectal cancer (CRC) patients and serves as a very useful model for studying adhesion-mediated mechanisms underlying CRC development [1][2]. This notion is supported by findings demonstrating that β-catenin plays a dual role in the cell. It is a major linker of cell–cell adhesion receptors (of the cadherin type) to the actin-cytoskeleton and, in addition, β-catenin plays a critical role in transmitting the Wnt signal to the nucleus by being a co-transcriptional activator [together with T-cell factor (TCF)] of Wnt target genes in the nucleus [3][4]. These two seemingly unrelated roles of β-catenin and the characteristic hyperactivation of Wnt/β-catenin signaling in CRC can serve as a useful system for investigating the roles of adhesion-mediated and Wnt signaling in CRC invasion and metastasis.
Wnt signaling was discovered over 40 years ago [5][6] and was first shown to play a role in determining the segmentation pattern in Drosophila [7]. Following these original studies, in the coming years, a role for Wnt signaling in embryonic axis determination in vertebrates was reported [8], and the potential involvement of the Wnt pathway in cancer development in humans was suggested [9]. In parallel, numerous studies addressed the identification of downstream components in the Wnt signaling pathway and discovered that inactivating mutations in the adenomatous polyposis coli (APC) gene, which is involved in β-catenin degradation, is a key step in the activation of Wnt signaling during CRC development [10]. In addition, stabilizing mutations in β-catenin against degradation by the ubiquitin-proteasomal system were also identified in a minority of CRC cases [11][12]. At this stage, an important avenue of research consisted of unraveling the target genes of Wnt/β-catenin signaling that are responsible for human CRC development. As the early steps in tumorigenesis are driven by changes that lead to uncontrolled proliferation of cells, initial studies focused on asking whether key regulators of the cell cycle (especially those leading to increased cell proliferation) are target genes of Wnt signaling and contain β-catenin/TCF binding sites in their promoter region. These studies led to the discovery of c-myc [13] and cyclin D1 [14][15] as target genes of β-catenin/TCF transactivation. Since then, hundreds of additional β-catenin-TCF target genes were discovered; for most of these genes, their role in CRC development remains to be determined [16]. Initial immunohistochemical studies of human CRC tissue did not detect a significant accumulation of β-catenin in the nuclei of early-stage CRC tissue and β-catenin localization remained mostly at cell–cell junctions in both normal colonic epithelial cells and in differentiated areas of CRC tissue [17][18]. However, at the later stages of CRC development, especially during the invasive and metastatic stages of tumor progression, a vast accumulation of β-catenin could be demonstrated, mostly in the nuclei of cancer cells [17][18], in addition to a specific expression of β-catenin target genes at the invasive areas of the tumor [19].
2. Members of the L1 Family of Cell Adhesion Receptors Are β-Catenin-TCF Target Genes
Initial DNA microarray analyses of genes induced by activated β-catenin-TCF signaling in cancer cells identified two members of the L1 family of immunoglobulin-like cell adhesion receptors, NrCAM [20][21] and L1 [19]. These findings were unexpected because both L1 and NrCAM were known to be present mostly in nerve cells, playing key roles during brain development by regulating a number of dynamic cellular processes including axonal growth, fasciculation, and pathfinding [22][23]. In previous studies, numerous point mutations were discovered in the L1 molecule that have severe consequences on brain development, leading to mental retardation by a group of syndromes known as L1 syndrome, MASA syndrome, and X-linked hydrocephalus [24][25][26][27][28].
L1 is a cell adhesion transmembrane receptor, believed to act mostly by homophilic interactions with L1 on the surface of neighboring cells. L1 belongs to the superfamily of immunoglobulin-like cell adhesion receptors, containing six Ig-like domains and five fibronectin type III repeats; a transmembrane sequence; and a highly conserved (from C. elegans to man) cytoplasmic tail that has binding sites for ezrin, ankyrin, and other PDZ-containing proteins (Figure 1). In addition, L1 can be cleaved in the juxtamembrane region, outside the cell, by the metalloprotease ADAM10, and inside the cell, it has binding sites for the γ-secretase cleavage complex (Figure 1). Unlike cadherins that are characterized by strong homophilic interactions, L1 can interact via both homophilic and heterophilic binding to other neuronal cell adhesion molecules including neurocan, neuropilin1, axonin, and N-CAM [29]. In addition, L1 can associate with ECM components (fibronectin, laminin, tenascin) and ECM receptors (integrins) and can also bind to growth factor receptors, such as EGFR and basic FGFR [30]. Because of these numerous weak interactions of L1 with a variety of molecules, an increase in the expression of L1 in cancer cells could be advantageous for promoting the motile, invasive, and metastatic stages of tumorigenesis.
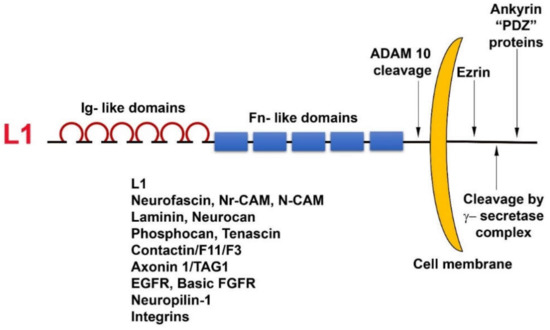
Figure 1. Domain structure and binding partners of L1. Note the numerous types of L1 ligands in the ectodomain as well as in the cytoplasmic tail domain of the molecule.
3. The Roles of L1 in Promoting CRC Cell Proliferation, Motility, Tumorigenesis, and Metastasis
Overexpression of L1 in 3T3 mouse fibroblasts and in human CRC cell lines results in elevated cell proliferation under stress (in the absence of serum); increased motility; invasion; tumorigenesis upon s.c injection into mice [19]; and, in the case of CRC cells, metastasis to the liver, a hallmark of human CRC progression [31]. The metalloprotease ADAM10, also a target gene of β-catenin-TCF transactivation, cleaves the ectodomain of L1 (Figure 1), thereby leading to its shedding, and promotes the rebinding of the shed L1 ectodomain to L1 molecules on the cell surface and enhances the metastatic potential of human CRC cells [31].
Immunohistochemical analysis of human CRC tissue revealed that the more differentiated areas of the tumor and the normal colonic epithelium do not express L1 [19]. L1 is exclusively expressed at the invading edge of human CRC tissue (Figure 2) in the membrane of cells that display strong nuclear β-catenin staining, indicative of a highly active β-catenin-TCF transactivation [19]. These results were recently confirmed and extended to show that, while L1 is not required for adenoma initiation, it plays multiple roles in cancer propagation, liver metastasis, and chemoresistance [32]. This study also demonstrated that L1 is not expressed in the homeostatic intestinal epithelium, but its expression is required for CRC organoid formation and metastasis initiation and growth. Finally, L1 expression was shown to be crucial for the regrowth occurring during wound healing in the intestine following injury [32]. Taken together, these results are reminiscent of the important roles played by L1 in the dynamic cellular processes occurring in nerve cells during brain development (i.e., axonal growth, pathfinding, and fasciculation) [33].
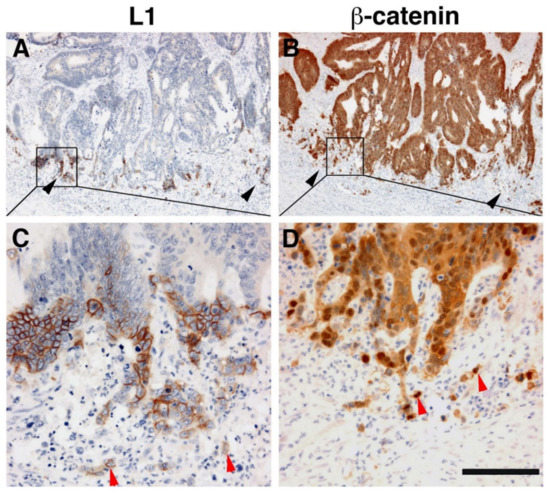
Figure 2. L1 is exclusively expressed at the invasive front of human colorectal cancer (CRC) tissue in cells expressing β-catenin in their nuclei. (A) Immunohistochemical staining of human CRC tissue for L1. Note the preferential localization of L1 in invasive areas of the tumor (black arrowheads), but not in the inner more differentiated areas of the tumor. (B) In contrast to L1 localization, a serial tissue section stained with anti β-catenin antibody displays a uniform staining of the same CRC tissue area. (C) Enlarged picture of the boxed area in (A) showing the membranal localization of L1. Single CRC cells invading into the stroma could also be seen (red arrowheads). (D) Magnified picture of the boxed area shown in (B) localizing β-catenin staining in both the cytoplasm and nuclei of CRC tissue cells and in the nuclei of single invasive cells (red arrowheads) at the tumor tissue edge [19]. Scale bar: (A,B) 375 μm, (C,D) 75 μm.
References
- Valenta, T.; Hausmann, G.; Basler, K. The many faces and functions of β-catenin. EMBO J. 2012, 31, 2714–2736.
- Basu, S.; Cheriyamundath, S.; Ben-Ze’Ev, A. Cell–cell adhesion: Linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Research 2018, 7, 1488.
- McCrea, P.D.; Gottardi, C.J. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 2016, 17, 55–64.
- Gumbiner, B.M. Cell Adhesion: The Molecular Basis of Tissue Architecture and Morphogenesis. Cell 1996, 84, 345–357.
- Sharma, R. Wingless, a new mutant in D. melanogaster. J. Dros. Inf. Service 1973, 50, 134.
- Sharma, R.; Chopra, V. Effect of the wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev. Biol. 1976, 48, 461–465.
- Nüsslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nat. Cell Biol. 1980, 287, 795–801.
- McMahon, A.P.; Moon, R.T. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell 1989, 58, 1075–1084.
- Nusse, R.; Varmus, H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 1982, 31, 99–109.
- Kinzler, K.W.; Vogelstein, B. Lessons from Hereditary Colorectal Cancer. Cell 1996, 87, 159–170.
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a beta -Catenin-Tcf Complex in APC-/- Colon Carcinoma. Science 1997, 275, 1784–1787.
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of beta -Catenin-Tcf Signaling in Colon Cancer by Mutations in beta -Catenin or APC. Science 1997, 275, 1787–1790.
- He, T.-C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a Target of the APC Pathway. Science 1998, 281, 1509–1512.
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’Ev, A. The cyclin D1 gene is a target of the -catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527.
- Tetsu, O.; McCormick, F. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nat. Cell Biol. 1999, 398, 422–426.
- Herbst, A.; Jurinovic, V.; Krebs, S.; Thieme, S.E.; Blum, H.; Göke, B.; Kolligs, F.T. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genom. 2014, 15, 74.
- Brabletz, T.; Jung, A.; Hermann, K.; Günther, K.; Hohenberger, W.; Kirchner, T. Nuclear Overexpression of the Oncoprotein β-Catenin in Colorectal Cancer is Localized Predominantly at the Invasion Front. Pathol.–Res. Pr. 1998, 194, 701–704.
- Brabletz, T.; Jung, A.; Reu, S.; Porzner, M.; Hlubek, F.; Kunz-Schughart, L.A.; Knuechel, R.; Kirchner, T. Variable -catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA 2001, 98, 10356–10361.
- Gavert, N.; Conacci-Sorrell, M.; Gast, D.; Schneider, A.; Altevogt, P.; Brabletz, T.; Ben-Ze’Ev, A. L1, a novel target of β-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J. Cell Biol. 2005, 168, 633–642.
- Conacci-Sorrell, M.E.; Ben-Yedidia, T.; Shtutman, M.; Feinstein, E.; Einat, P.; Ben-Ze’Ev, A. Nr-CAM is a target gene of the beta -catenin/LEF-1 pathway in melanoma and colon cancer and its expression enhances motility and confers tumorigenesis. Genes Dev. 2002, 16, 2058–2072.
- Bottomly, D.; Kyler, S.L.; McWeeney, S.K.; Yochum, G.S. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010, 38, 5735–5745.
- Zelina, P.; Avci, H.X.; Thelen, K.; Pollerberg, G.E. The cell adhesion molecule NrCAM is crucial for growth cone behaviour and pathfinding of retinal ganglion cell axons. Development 2005, 132, 3609–3618.
- Sakurai, T. The role of NrCAM in neural development and disorders—Beyond a simple glue in the brain. Mol. Cell. Neurosci. 2012, 49, 351–363.
- Jouet, M.; Rosenthal, A.; Armstrong, G.; MacFarlane, J.; Stevenson, R.; Paterson, J.; Metzenberg, A.; Ionasescu, V.; Temple, K.; Kenwrick, S. X–linked spastic paraplegia (SPG1), MASA syndrome and X–linked hydrocephalus result from mutations in the L1 gene. Nat. Genet. 1994, 7, 402–407.
- Wong, E.V.; Kenwrick, S.; Willems, P.; Lemmon, V.P. Mutations in the cell adhesion molecule LI cause mental retardation. Trends Neurosci. 1995, 18, 168–172.
- Fransen, E.; van Camp, G.; Vits, L.; Willems, P.J. L1-associated diseases: Clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 1997, 6, 1625–1632.
- Kamiguchi, H.; Hlavin, M.L.; Lemmon, V.P. Role of L1 in Neural Development: What the Knockouts Tell Us. Mol. Cell. Neurosci. 1998, 12, 48–55.
- Kenwrick, S.; Watkins, A.; de Angelis, E. Neural cell recognition molecule L1: Relating biological complexity to human disease mutations. Hum. Mol. Genet. 2000, 9, 879–886.
- Altevogt, P.; Doberstein, K.; Fogel, M. L1CAM in human cancer. Int. J. Cancer 2015, 138, 1565–1576.
- Schmid, R.S.; Maness, P.F. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 2008, 18, 245–250.
- Gavert, N.; Sheffer, M.; Raveh, S.; Spaderna, S.; Shtutman, M.S.; Brabletz, T.; Barany, F.; Paty, P.; Notterman, D.; Domany, E.; et al. Expression of L1-CAM and ADAM10 in Human Colon Cancer Cells Induces Metastasis. Cancer Res. 2007, 67, 7703–7712.
- Ganesh, K.; Basnet, H.; Kaygusuz, Y.; Laughney, A.M.; He, L.; Sharma, R.; O’Rourke, K.P.; Reuter, V.P.; Huang, Y.-H.; Turkekul, M.; et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Rev. Cancer 2020, 1, 28–45.
- Wiencken-Barger, A.; Mavity-Hudson, J.; Bartsch, U.; Schachner, M.; Casagrande, V.A. The Role of L1 in Axon Pathfinding and Fasciculation. Cereb. Cortex 2004, 14, 121–131.




