
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Munizay Paracha | + 4729 word(s) | 4729 | 2021-02-18 09:00:34 | | | |
| 2 | Camila Xu | Meta information modification | 4729 | 2021-03-29 03:52:45 | | |
Video Upload Options
Microsatellite instability (MSI) is a molecular marker of deficient DNA mismatch repair (dMMR) that is found in approximately 15% of colorectal cancer (CRC) patients.
1. Introduction
Colorectal cancer (CRC) is the third most common and second most deadly cancer worldwide, causing an estimated 880,000 deaths in 2018 [1]. Mortality rates for CRC have been declining in many countries due to improved screening efforts and therapeutic advances [2], but both the incidence and mortality of CRC have been increasing in patients under the age of 50 in high-income countries [3]. CRC is a heterogenous group of diseases (subtypes) with differences in epidemiology, anatomy, histology, genomics, transcriptomics and host immune response [4][5][6][7]. This heterogeneity leads to disparate clinical presentation, survival and response to therapy [8][9][10][11].
One of the clinically relevant subtypes of CRC is DNA mismatch repair deficient (dMMR) CRC. dMMR occurs due to pathogenic alterations in genes involved in the MMR system (MLH1, MSH2/EPCAM, MSH6, and PMS2) [12][13][14]. Several mechanisms can lead to dMMR, the most common being somatic hypermethylation of the MLH1 gene promoter [14]. In patients with Lynch Syndrome, who carry a germline pathogenic mutation in one of the MMR genes, an additional somatic alteration can occur (“second hit”), leading to the phenotype of dMMR tumors. Sporadic bi-allelic somatic mutations in the MMR genes can also occur [13]. Deficiencies in MMR cause high rates of mutations throughout the DNA, especially in microsatellites, regions of DNA in which short sequences of nucleotides are repeated in tandem [12]. Thus, dMMR results in microsatellite instability (MSI), which is a highly sensitive marker of dMMR [13].
MSI has diagnostic, prognostic, and therapeutic implications in CRC and other cancers. The detection of dMMR/MSI is recommended as a screening test for Lynch syndrome for every case of CRC [14]. Confirmation requires germline testing and can inform surveillance and treatment decisions for both the patient and their relatives [15]. CRC patients with MSI generally have a better prognosis [13][16][17], which may be explained by a robust host immune response to tumor neoantigens [10][11][18]. Lastly, MSI status can inform treatment decisions, as patients with MSI tumors may be eligible for immune checkpoint inhibitor (ICI) therapy [19], and the benefit of fluorouracil-based chemotherapy regimens for tumors with MSI has been questioned [20][21][22][23].
Current testing for dMMR/MSI requires either an immunohistochemical analysis of MMR protein expression or a PCR-based assay of microsatellite markers [14]. While guidelines set forth by multiple professional societies recommend universal testing for dMMR/MSI [24], these methods require additional resources and are not available at all medical facilities, so many CRC patients are not currently tested [25]. Recently, artificial intelligence has been evaluated as a method to predict MSI directly from hematoxylin and eosin (H&E) stained slides (Figure 1). If successful, this approach could have significant benefits, including reducing cost and resource-utilization and increasing the proportion of CRC patients that are tested for MSI.
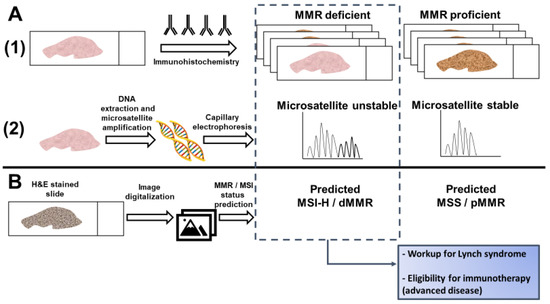
Figure 1. Detection of microsatellite instability (MSI) or mismatch repair (MMR) deficiency is performed by (A1) Immunohistochemistry of the mismatch repair proteins or (A2) PCR amplification of consensus microsatellite repeats that are analyzed with capillary electrophoresis. Inference of MSI/MMR status from next generation sequencing (NGS) is not presented. (B) MSI/MMR status can be predicted from hematoxylin and eosin (H&E) stained slides, without requiring molecular analyses (see Figure 2). Detection of MSI/dMMR has implications for Lynch Syndrome screening and determining eligibility for immune checkpoint blockade in advanced disease. MSS: microsatellite stable. MSI-H: high microsatellite instability. pMMR: proficient mismatch repair. dMMR: deficient mismatch repair.
2. Histological and Clinical Predictors of Microsatellite Instability
With the significant cost and non-universal availability of the molecular testing required to determine MMR/MSI status, studies have sought to predict MSI based on routinely available data, such as clinical information and histopathology [26]. CRC tumors with MSI are associated with certain histological features, detectable via standard H&E staining, and clinical data, such as patient age and tumor location [26][27][28]. Similar observations have been made in other tumors enriched for MSI, such as endometrial cancer [29]. These associations may present a means of identifying the tumors most likely to have the dMMR phenotype, and therefore the patients most likely to benefit from additional testing. They may also help to identify those at low risk who would be less likely to benefit. The targeted deployment of MMR/MSI testing could reduce costs and save resources [26]. Inferring MSI status may be considered in settings where MSI testing is not performed but is unlikely to be adopted in resource-rich settings unless the prediction accuracy is near-perfect.
Several clinicopathologic predictors of MSI have been discovered and several groups have proposed models for MSI prediction (Table 1). Histological features such as signet ring cells, mucinous or medullary morphology, and poor differentiation are significantly associated with MSI status, but show poor sensitivity for MSI prediction on their own [27][30]. Correlations between MSI and immunological features of tumor pathology, such as measurements of tumor infiltrating lymphocytes (TILs) [11][26][28][31] and specific histological structures such as the Crohn’s-like lymphoid reaction (CLR), are well established in the literature [18][26][28]. CLR represents CRC-specific tertiary lymphoid aggregates [18]. The host response to MSI tumors is attributed to the high tumor mutational burden (TMB) and the abundance of immunogenic mutations, including insertion-deletion mutations, but other factors may contribute [32][33][34]. The Revised Bethesda Guidelines for MSI testing in CRC suggested testing tumors with “MSI histology” in patients younger than 60 years of age [35]. MSI histology was defined as the presence of TILs, CLR, mucinous/signet-ring differentiation, or medullary growth pattern. One of the histopathological features most strongly associated with MSI is the density of TILs [26][27][30]. When TIL density was assessed as a potential predictor of MSI, the area under the receiver operating characteristic curve (AUC) was 0.73. With a cutoff value of 40 lymphocytes/0.94 mm2, MSI status could be predicted with a sensitivity of 75% and a specificity of 67% [30]. However, given that TIL density can vary across tumor area, this study using surgical specimens likely yielded a greater AUC than would be achieved with smaller biopsy specimens, such as those typically available from sites of metastasis.
Table 1. Histological predictors of microsatellite instability.
| A | |||||
| Variable | Sensitivity % (95% CI) | Specificity % (95% CI) | Odds Ratio, Univariate (95% CI) | Odds Ratio, Multivariate (95% CI) | |
| Host response features | |||||
| Tumor-infiltrating lymphocytes (TILs) | 70 [26] | 76 [26] | 7.4 (5.4–10.3) [26] | 3.8 (2.5–5.6) [26] | |
| 72 (64–78) [27] | 82 (80–85) [27] | 9.1 (5.9–14.1) [27] | |||
| 21 [30] | 97 [30] | 9.8 (3.5–28.5) [30] | |||
| 60 (50–70) [36] | 78 (76–79) [36] | 5.2 (3.2–8.5) [36] | 3.7 (2.0–6.8) [36] | ||
| Crohn’s-like lymphocytic reaction (CLR) | 68 [26] | 54 [26] | 2.5 (1.8–3.4) [26] | 2.3 (1.6–3.5) [26] | |
| 56 (48–63) [27] | 77 (74–80) [27] | 1.9 (1.2–2.9) [27] | |||
| 49 [30] | 64 [30] | 1.7 (0.9–3.2) [30] | |||
| 69 (59–78) [36] | 45 (44–46) [36] | 1.8 (1.1–3.0) [36] | 1.1 (0.6–2.0) [36] | ||
| Peritumoral lymphocytic reaction | 86 (77–92) [36] | 42 (40–42) [36] | 4.3 (2.3–8.3) [36] | 3.7 (1.6–8.6) [36] | |
| Stromal plasma cells | 78 (68–86) [36] | 48 (46–48) [36] | 3.2 (1.9–5.6) [36] | 2.1 (1.1–4.1) [36] | |
| Tumor characteristics | |||||
| Mucinous morphology * | 53 [26] | 80 [26] | 4.6 (3.4–6.3) [26] | 1.7 (1.1–2.7) [26] | |
| 28 (21–35) [27] ** | 91 (89–93) [27] ** | 2.8 (1.7–4.8) [27] ** | |||
| 22 [30] | 93 [30] | 3.7 (1.7–8.0) [30] | |||
| 51 (41–61) [36] | 78 (77–79) [36] | 3.7 (2.3–6.0) [36] | 4.71 (2.1–10.7) [36] | ||
| 2.13 (1.3–3.4) [37] | |||||
| Medullary morphology (10–70%) | 25 [30] | 97 [30] | 12.5 (4.6–35.9) [30] | ||
| Grade † | 64 [26] † | 81 [26] † | 7.4 (5.4–10.1) [26] † | 3.4 (2.2–5.2) [26] † | |
| 38 (31–46) [27] | 82 (79–84) [27] | 1.9 (1.2–3.1) [27] | |||
| 38 [30] | 87 [30] | 4.0 (2.2–7.3) [30] | |||
| 17 (10–26) [36] | 90 (89–91) [36] | 1.8 (0.9–3.5) [36] | |||
| 32 (23–43) [36] † | 77 (76–78) [36] † | 1.6 (0.9–2.6) [36] † | |||
| Signet ring cells | 4.3 (2.2–8.7) [26] | ||||
| 13 [30] | 95 [30] | 2.7 (1.1–6.8) [30] | |||
| Lack of dirty or garland necrosis | 59 [26] | 79 [26] | 5.4 (3.9–7.4) [26] | 1.8 (1.1–2.8) [26] | |
| 26 (18–35) [36] | 89 (88–90) [36] | 2.7 (1.5–4.7) [36] | 1.4 (0.7–3.0) [36] | ||
| Cribriform pattern | 13 [30] | 72 [30] | 0.4 (0.2–0.8) [30] | ||
| Histologic heterogeneity | 4.4 (3.0–6.4) [26] | ||||
| 55 (45–65) [36] | 69 (68–70) [36] | 2.7 (1.7–4.4) [36] | |||
| Clinical/Molecular Features | |||||
| Age <50 years | 2.2 (1.3–3.8) [26] | 3.1 (1.5–6.5) [26] | |||
| 52 (44–60) [27] | 59 (56–62) [27] | 1.9 (1.3–2.9) [27] | |||
| 21 (13–29) [36] | 89 (88–90) [36] | 2.0 (1.1–3.7) [36] | 3.8 (1.8–8.0) [36] | ||
| Female | 1.4 (1.0–1.9) [26] | ||||
| 51 (41–62) [36] | 63 (62–64) [36] | 1.8 (1.1–2.8) [36] | 1.3 (0.7–2.2) [36] | ||
| 1.56 (1.0–2.4) [37] | |||||
| Size > or equal to 60 mm | 2.75 (1.8–4.2) [37] | ||||
| Anatomic site (right sided/proximal) | 70 [26] | 63 [26] | 4.1 (2.9–5.7) [26] | 2.2 (1.5–3.3) [26] | |
| 74 (67–81) [27] | 70 (67–73) [27] | 4.7 (3.1–7.3) [27] | |||
| 79 (70–87) [36] | 63 (61–63) [36] | 6.4 (3.6–11.2) [36] | 5.08 (2.7–9.6) [36] | ||
| 3.76 (2.4–5.9) [37] | |||||
| BRAF mutant | 13.33 (8.0–22.2) [37] | ||||
| B | |||||
| Model | Model Variables | Sensitivity (%) | Specificity (%) | Positive/Negative Predictive Value (%) | AUC or Accuracy (95% CI) |
| Greenson et al. ** [26] | TIL/HPF, well or poorly differentiated, age < 50, CLR, R-sided, lack of dirty necrosis, any mucinous differentiation | 92 ** | 46 ** | AUC 0.850 ** | |
| MsPath [27] | Age < 50, proximal location, mucinous/signet ring/undifferentiated, poorly differentiated, CLR, TILs | 93 | 55 | AUC 0.890 (0.83– 0.94) | |
| PREDICT [36] | R-sided, mucinous component, age < 50 years, TILs, peritumoral reaction, increased stromal plasma cells | 96.9 | 76.6 | 35.2/99.5 | AUC 0.924 |
| Fujiyoshi et al. [37] | Female, mucinous component, tumor size > or equal to 60 mm, proximal location, BRAF mutation | 76 | 77 | AUC 0.856 (0.806 –0.905) | |
| RERTest6 ** [38] | Proximal location, expansive growth pattern, CLR, solid pattern %, mucinous pattern %, cribriform pattern | 78.01 ** | 93.39 ** | 51.8/97.9 ** | Accuracy 0.921 ** |
Table 1 summarizes individual features and composite models used for the prediction of microsatellite instability (MSI) from histology. (A) Individual variables associated with MSI are listed. * Jenkins et al. [27] included signet cell and medullary morphology with mucinous morphology. † These studies included either poorly or well differentiated tumors as associated with MSI. (B) Multivariable models and their performance in differentiating between tumors with and without MSI are presented. ** Values are presented for validation cohorts when available. Greenson et al. and RERTest6 did not include a separate validation cohort. CI: confidence interval. TIL: tumor infiltrating lymphocytes. CLR: Crohn’s-like lymphoid reaction. HPF: high power field. R-sided: right sided. AUC: area under the curve.
Multiple histological and clinical variables have been incorporated into algorithms designed to predict MSI status. The MsPath score was developed to predict MSI in patients under the age of 60 [27]. Using a scoring system incorporating age, anatomical site of the primary tumor, histologic type, tumor grade, and the presence or absence of TILs and CLR, an AUC of 0.89 was achieved when the model was tested against a separate validation cohort (Table 1). Validation of the MsPath score in a population based-cohort showed that its accuracy was insufficient for the selection of patients for Lynch Syndrome germline testing, misclassifying 18% (2/11) of patients with a pathogenic mutation in MLH1/MSH2 [39]. Another scoring scheme by Greenson et al. incorporated similar variables but included lack of dirty necrosis in the model and was derived from a population that included patients of all ages [26]. The features associated with MSI all had a negative predictive value >90%. This model yielded an AUC of 0.85 based on the study cohort alone (no validation cohort was tested) (Table 1). Over half of the tumors analyzed had less than 5% chance of harboring MSI, presenting the potential for significant cost savings [26]. In another cohort, the model by Greenson et al. detected 93% of tumors with MSI and outperformed MsPath [40].
The PREDICT score was developed to improve on MsPath and other models [36]. It included variables that were significantly associated with MSI in a multivariable regression model, including age <50, right sided location, TILs, a peritumoral lymphocytic reaction, any mucinous component and increased stromal plasma cells [36]. PREDICT reported a sensitivity of 97% for the detection of MSI with an AUC of 0.924 in the validation cohort (Table 1). The RERtest6 model was developed to maximize the negative predictive value and included tumor location, growth pattern, solid and mucinous pattern, TIL and CLR [38]. The model had an accuracy of 92% in the global cohort and a negative predictive value of 97.9% (Table 1). The prevalence of MSI was 8.5% in this study. If this model were applied as screening for MSI in this study population, only 10% of patients would need confirmatory testing [38].
Another large study of MSI prediction from commonly available clinico-pathologic data included over three thousand patients over 50 years of age in Japan [37]. Female sex, proximal location, tumor size larger than 60 mm, mucinous component and BRAF mutation were associated with MSI and were included in a composite score used for prediction. CLR and TILs were not evaluated. In the validation cohort, the AUC was 0.856. Patients with MLH1 promoter hypermethylation had higher scores than patients with Lynch Syndrome, as a result of the known association between BRAF mutations and MLH1 hypermethylation and the high score given to BRAF mutations in the model. Overall, the performance of the model was disappointing, with approximately 25% of MSI tumors misclassified at the proposed threshold [37].
The encouraging performance of certain histology-based prediction models has not been sufficient to supersede universal testing for MSI/dMMR. Measurement of the variables for MSI prediction requires significant effort and expertise by pathologists, and inter-rater differences may affect the perceived reliability of histology-based scoring systems [41][42]. However, this work is fundamental to the premise that MSI can be predicted from histology, which has now been proposed as a task for deep learning from digital pathology [43] (Figure 1).
3. Predicting MSI Status with Deep Learning
Recently, several studies have investigated the potential for CNNs to predict MSI from H&E stained histological samples. Kather et al. trained and tested CNNs on gastric, endometrial, and colorectal samples that were snap-frozen or formalin-fixed paraffin-embedded (FFPE) [43]. FFPE slides are routinely used for histological diagnosis and immunohistochemistry. Fixation with formalin and embedding with paraffin are performed to maintain tissue architecture and morphology, and to allow long-term preservation at room temperature. The process of generating an FFPE slide requires many hours and the fixation process results in the cross-linking of DNA and proteins that can impair the performance of molecular analyses. Snap-frozen tissue is not routinely obtained but can be used for intraoperative diagnoses because it can be rapidly reviewed by a pathologist. Snap-frozen tissue can also be used for extensive molecular analyses [44][45]. The morphological quality of snap-frozen tissue is not considered sufficient to render a definitive diagnosis, and confirmation using FFPE slides is typically required [46][47][48]. All CNNs in the study by Kather et al. had been pretrained on the ImageNet database, and only the last ten layers of the CNNs were trainable. After assessing the performance of five CNNs in differentiating tumor tissue from healthy tissue, the CNN ResNet-18 (a ResNet with 18 layers) was selected for further evaluation based on its strong performance and smaller number of parameters. The advantage of a CNN with a smaller number of parameters is a decreased risk of overfitting the data and increased likelihood of maintaining performance when applied to a validation cohort. ResNet-18 was trained with two sets of CRC (fresh frozen and FFPE slides) and one gastric cancer dataset (FFPE) from TCGA (Table 2). Tumor tissue was divided into smaller tiles, each of which was separately analyzed and assigned a predicted MSI score. Predicted MSI status for each slide was determined by the predicted MSI status of the majority of its constituent tiles.
Table 2. Deep learning for prediction of MSI from digital pathology.
| CNN and Additional Methods | Other CNNs Evaluated | Training Cohort | Test Cohort(s) with AUC (95% CI) or Accuracy | External Validation Cohort(s) with AUC (95% CI) |
|---|---|---|---|---|
| ResNet-18 Whole-slide image classified per majority of image tiles [43] |
AlexNet, VGG-19, InceptionV3, SqueezeNet | TCGA CRC FFPE | TGCA CRC FFPEAUC 0.77 (0.62–0.87) | DACHS CRC FFPE AUC 0.84 (0.72–0.92) |
| TCGA CRC frozen | TCGA CRC frozen AUC 0.84 (0.73–0.91) |
DACHS CRC FFPE 0.61 (0.50–0.73) |
||
| TCGA gastric FFPE | TCGA gastric FFPE AUC 0.81 (0.69–0.90) |
DACHS CRC FFPE AUC 0.60 (0.48–0.69) KCCH gastric FFPE AUC 0.69 (0.52–0.82) |
||
| TCGA uterine FFPE | TCGA uterine FFPE AUC 0.75 (0.63–0.83) |
|||
| ShuffleNet [49] | AlexNet, InceptionV3, ResNet-18, DenseNet201 | TCGA CRC | TCGA CRC AUC 0.805 |
DACHS CRC AUC 0.89 (0.88–0.92) |
| ResNet-18 Whole slide image classified using two multiple instance learning pipelines integrated into an ensemble classifier [50] |
none | TCGA CRC frozen | TCGA CRC frozen AUC 0.885 |
Asian CRC AUC FFPE 0.650 |
| TCGA CRC frozen with 10% Asian CRC FFPE | Asian CRC FFPEAUC 0.850 | |||
| TCGA CRC frozen with 70% Asian CRC FFPE | Asian CRC FFPEAUC 0.926 | |||
| Custom multilayer perceptron (HE2RNA) applied after feature extraction by ResNet-50; with and without transcriptomic representation of histology [51] | none | TCGA CRC FFPE, with transcriptomic representation and 20% of training cohort | TCGA CRC FFPEAUC ~0.80 * | |
| TCGA CRC FFPE, using >80% of training cohort | TCGA CRC FFPEAUC ~0.80 * | |||
| Inception-V3 with and without adversarial learning [52] | VGG-19, ResNet-50 | TCGA CRC | TCGA CRC Accuracy 98.3% |
TCGA endometrial Accuracy 53.7% |
| TCGA CRC and endometrial | TCGA CRC Accuracy 72.3% TCGA endometrial Accuracy 84.2% |
TCGA gastric Accuracy 34.9% |
||
| TCGA CRC and endometrial with adversarial learning | TCGA CRC Accuracy 85.0%TCGA endometrial Accuracy 94.6% |
TCGA gastric Accuracy 57.4% |
||
| InceptionResNetV1 [53] | InceptionV1-3, InceptionResnetV1-2, Panoptes1-4 (multibranch custom InceptionResnet) | TCGA and CPTAC endometrial carcinoma | TCGA and CPTAC endometrial carcinoma AUC 0.827 (0.705–0.948) |
|
| ShuffleNet [54] | none | MSIDETECT CRC (color normalized) | MSIDETECT CRC AUC 0.92 (0.90–0.93) |
YCR-BCIP CRC surgical samples AUC 0.96 (0.93–0.98) |
| YCR-BCIP CRC biopsy samples AUC 0.78 (0.75–0.81) |
||||
| YCR-BCIP CRC biopsy samples | YCR-BCIP CRC biopsy samples |
Table 2 summarizes the methods and performance of available data regarding deep learning for MSI prediction from pathology. * Exact AUC not reported, estimate based on an included graphical representation. CNN: convolutional neural network. TCGA: The Cancer Genome Atlas, CRC: colorectal cancer, DACHS: CRC prevention through screening study (abbreviation in German), FFPE: Formalin-Fixed Paraffin-Embedded, AUC: Area under the receiver operating characteristic curve, KCCH: Kanagawa Cancer Center Hospital (Japan), CPTAC: Clinical Proteomic Tumor Analysis Consortium, MSIDETECT: A consortium comprised of whole slide images from TCGA, DACHS, the United Kingdom-based Quick and Simple and Reliable trial (QUASAR), and the Netherlands Cohort Study (NLCS), YCR-BCIP: Yorkshire Cancer Research Bowel Cancer Improvement
Using this process, the CNN was able to detect MSI in snap-frozen and FFPE TCGA samples with similar AUCs to those achieved with previous pathology-based scoring systems such as MsPath and the model by Greenson et al. (0.84 for snap-frozen CRC samples, 0.77 for FFPE CRC samples, and 0.81 in FFPE gastric adenocarcinoma samples). This level of performance was maintained when the CNN trained on FFPE CRC samples was tested on an external validation cohort from the DACHS (Darmkrebs: Chancen der Verhütung durch Screening) study (Table 2), which consisted of FFPE CRC samples from Germany (AUC 0.84). The authors also tested the classification performance of the ResNet when applied to slides with limited tissue, finding that performance plateaued with a quantity of tissue that is available from standard needle biopsies [43].
To attempt to identify what pathological features the ResNet used to make its classifications, tumor regions that were assigned high or low MSI scores were visually inspected. Areas predicted by the CNN to represent MSI often showed characteristics consistent with known pathological correlates of MSI, such as poor differentiation and lymphocytic infiltration. PD-L1 expression and an interferon gamma transcriptomic signature were correlated with the proportion of a sample’s tiles predicted to have MSI. This finding is consistent with previous data showing high expression of PD-L1 and interferon gamma in CRC with MSI [55][56].
Despite encouraging performance for MSI classification in similar cohorts, testing against different cohorts revealed some limitations. CNNs trained on snap-frozen CRC samples or gastric adenocarcinoma samples did not perform as well as the CNN both trained and tested on FFPE CRC samples. When the CNN was trained to detect MSI in endometrial cancers, its performance was significantly reduced to an AUC of 0.75, raising the possibility that the CNN is learning tissue-specific features associated with MSI. Additionally, the CNN trained on TCGA gastric adenocarcinomas did not perform as well when tested on a Japanese gastric adenocarcinoma cohort (AUC 0.69), possibly due to distinctive histological patterns seen in gastric adenocarcinomas in this cohort [43].
Other studies have attempted to improve upon these results using other CNNs and machine learning techniques (Table 2). In a follow up study by Kather et al., the prediction of MSI was performed as a benchmark task by various CNNs, which were pretrained on the ImageNet database [49]. The ResNet and Inception CNNs were outperformed by the DenseNet [57] and ShuffleNet [58] architectures. ShuffleNet, a CNN optimized for mobile devices, was able to achieve an AUC of 0.89 when trained on a CRC cohort from TCGA and validated on the DACHS CRC cohort (Table 2). The ResNet used for the previous study by Kather et al. achieved an AUC of 0.84 [43][49].
Another group reports improvement upon the results by Kather et al. in terms of overall predictive accuracy and generalizability to different cohorts [50]. This study also used ResNet-18 to assign each tile within the tumor area an MSI likelihood. However, multiple instance learning was used to train the CNN to classify the whole slide image. Multiple instance learning assumes that not all tumor regions contribute the same amount of information to the task of classification of the tumor as a whole [59]. Certain regions or patterns found in limited areas of a sample may be more important to determining the likelihood of the tumor being MSI. For example, any mucinous differentiation increases the likelihood of a tumor harboring dMMR/MSI [26][28]; this may be focal and not seen in the majority of tumor areas. Two different multiple instance learning methods were used in this study, and their input was integrated into a final ensemble predictor (Table 2). This ensemble classifier achieved an AUC of 0.885 [50], which was better than the performance reported by Kather et al. [43].
This group also found a significant reduction in AUC (0.650) when the TCGA-trained ensemble classifier was tested on a cohort of Asian patients with samples acquired with a different slide preparation protocol [50]. They were able to overcome this reduction in performance by transfer learning. By adding increasing proportions of data from the Asian cohort to the training set, they were able to achieve an AUC of 0.850 with 10% samples from the Asian cohort, with continued improvement up to an AUC of 0.926 with 70% samples from the Asian cohort (Table 2) [50]. Pathologic signatures were derived from the model and were associated with known features of MSI, including TMB and insertion-deletion mutational burden, as well as transcription signatures of immune activation.
A conference paper by Wang et al. also assessed an alternative technique, Patch Likelihood Histogram (PALHI), for integrating tile-level MSI predictions into patient-level predictions using whole slide images from a TCGA endometrial cancer cohort [60]. First, a ResNet-18 pre-trained on ImageNet was trained to predict MSI for individual tiles on a subset of the TCGA cohort. PALHI then generated a histogram of the patch-level estimated MSI likelihoods, which were used to train a machine learning classifier called XGBoost to make patient-level predictions. The performance of a pipeline using PALHI to make patient-level predictions was compared to pipelines using another machine learning method, Bag of Words (BoW) and the “majority voting” method, using another subset of the TCGA cohort as a testing set. The three methods were each trained on both patches assigned binary “hard labels” and patches assigned “soft labels,” or MSI probabilities. The PALHI method trained using “soft labels” yielded the best performance on the test set, with an AUC of 0.75. By comparison, the AUCs for BoW and the majority method using “soft labels” were 0.71 and 0.56, respectively [60].
Transcriptomic prediction from H&E slides has also been used to improve MSI prediction when limited training data are available [51]. First, features were extracted from each tissue tile using the ResNet-50, pretrained on the ImageNet database. These features served as the input for a custom multilayer perceptron, which was trained to predict gene expression from RNA-Seq data. Multilayer perceptrons are neural networks composed of fully connected layers, typically without convolutional layers. This neural network was trained on pan-cancer and tissue-specific TCGA datasets and was able to predict several expression signatures, including adaptive immune response signatures [51]. For MSI prediction, the authors simulated a situation where a limited number of training slides are available at two sites. They showed that, using the transcriptomic representation trained at one site, they could improve MSI prediction at the second site. However, when increasing proportions of data at the second site were used for MSI prediction without integrating transcriptomic representation, this advantage was largely lost. Neither method achieved an AUC > 0.85 and no external validation set was used (Table 2) [51]. It is unclear if this approach would be applicable in real-life settings.
In a conference paper submitted to the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018) [52] and a related patent [61], adversarial learning was used to improve the generalizability of CNNs for MSI prediction across different cancers. The Inception-V3, ResNet-50 and VGG-19 CNNs were compared; Inception-V3 was chosen for downstream analysis. TCGA samples were used for both testing and training; this study did not use an external validation dataset. MSI status was categorized as stable, low instability or high instability. Inception-V3 was trained on CRC samples and achieved a slide-level accuracy of 98.3% with an internal validation set of 10% of TCGA slides. It is unclear if this level of accuracy represents overfitting. Accuracy was poor when applied to endometrial carcinoma samples at 54%, whereas training the CNN on both CRC and endometrial carcinoma decreased the accuracy of MSI prediction for CRC to 72% (Table 2). This CNN also performed poorly at classifying MSI in gastric adenocarcinoma with a slide-level accuracy of 35%. Next, a tumor type classifier was added to the CNN with an adversarial objective—to decrease the ability of the model to predict tumor type. The rationale for creating this adversarial objective is to remove tissue-specific features that are learned by the CNN, such that the model will recognize the features associated with MSI better. Adversarial training improved MSI classification across the three cancer types, but accuracy remained poor for gastric adenocarcinoma at 57% [52].
Focusing on endometrial cancer, a recent study available as a preprint generated CNNs that had three branches of an InceptionResNet architecture, each analyzing tiles at a different resolution [53]. An optional fully connected layer incorporating clinical features was also evaluated as a fourth branch. This structure, termed Panoptes, allowed the model to take into account both tissue-level and cellular-level structures, as would a human pathologist using a microscope. MSI classification was one of several tasks that the CNNs were trained to do. While the complex architecture showed strong performance in predicting many histological and molecular features, MSI was best predicted by the existing InceptionResnetV1 architecture, with an AUC of 0.827 (Table 2), which outperformed Kather’s previously described ResNet-18 architecture (AUC 0.75). The inclusion of clinical data did not seem to improve the model’s performance: when the age and BMI of the patient were added into the model, its performance did not significantly improve [53]. Predicted MSI was correlated with certain histological features, including intratumoral and peritumoral lymphocytic infiltrates.
The strongest-performing model for MSI prediction was developed by Echle et al. by training a CNN on a large cohort of H&E-stained CRC samples from the MSIDETECT consortium, which is comprised of whole slide images from TCGA, DACHS, the United Kingdom-based Quick and Simple and Reliable trial (QUASAR), and the Netherlands Cohort Study (NLCS) [54]. A modified version of the CNN ShuffleNet that was pre-trained on ImageNet was trained on whole slide images from MSIDETECT with known MSI or dMMR status and externally validated on a separate population-based cohort, Yorkshire Cancer Research Bowel Cancer Improvement Programme (YCR-BCIP). For each slide, tumor tissue was manually outlined and the slide was divided into smaller tiles. The patient-level prediction of MSI/dMMR was based on the average tile-level prediction for each patient. The CNN was first trained and tested on individual sub-cohorts. As in earlier-described studies [43][50][52], when a CNN trained on a single sub-cohort was tested on another sub-cohort, performance usually suffered. A positive correlation between the size of the training cohort and the performance of the model was noted. The CNN was then trained on increasing numbers of patients randomly selected from the MSIDETECT cohort. The model showed better performance with greater numbers of patients up until about 5000 patients, after which performance plateaued. After training with samples from 5500 patients, the model attained an AUC of 0.92 when tested on a separate set of patients from MSIDETECT. When tested on the external validation cohort (YCR-BCIP), the model attained a similarly impressive AUC of 0.95. Additionally, when slides were subjected to color normalization, the specificity at given levels of sensitivity increased and a slight improvement in AUC to 0.96 was demonstrated [54]. Though these results are encouraging, it is worth noting that the samples used to train and test this model were derived mostly from European patients. Further validation with more diverse cohorts and prospective studies will be necessary before this model can be applied in a broad clinical context.
Subgroup analysis did reveal some variation in the model’s performance for certain tumor characteristics. While the performance was consistent for tumors at stages I-III (AUCs 0.91–0.93), the AUC for stage IV tumors was lower (0.83). The authors do not discuss potential explanations for this discrepancy, but there was a similar reduction in AUC for tumors with high histologic grade (AUC for high grade tumors was 0.83). The relatively low prevalence of MSI/dMMR in stage 4 colorectal cancers would have decreased the number of available images from this subgroup available for training, as would the fact that stage 4 tumors are more likely to come from biopsy specimens than complete resection samples. This lower performance for stage 4 tumors is unfortunate given that ICI therapy is currently primarily used in late-stage colorectal cancer, reducing the model’s potential utility for guiding treatment decisions. Additionally, the model predicted MSI more effectively for colon cancer (AUC 0.91) than for rectal cancer (AUC 0.83). Performance did not vary significantly by tumor molecular characteristics (e.g., mutation status) [54].
As noted above, a previous study demonstrated that the performance of ResNet-18 in classifying MSI status plateaued with a quantity of tissue that can be obtained by needle biopsy [43]. However, Echle et al. found a significant decrease in AUC when the CNN trained on surgical specimens was tested on YCR-BCIP biopsy specimens as compared to YCR-BCP surgical specimens (0.78 vs. 0.96). Though size of specimen may be a factor here, artifacts from specimen acquisition and the fact that samples were derived only from luminal tumor tissue may also affect performance. When the authors performed a 3-fold cross-validated experiment using YCR-BCIP biopsy specimens to both train and test, the AUC improved to 0.89 [54]. However, the model was not tested on samples from sites of metastasis, which are commonly biopsied in the clinical setting. Thus, machine learning models may be effective in classifying the MSI status of biopsy specimens, but will likely perform best when trained on similarly derived specimens.
Taken together, these studies demonstrate that multiple CNNs and machine learning techniques are being evaluated for MSI prediction from histology. There is no clear consensus regarding the optimal network architecture. The use of large and diverse datasets for training may overcome some of the limitations of models whose classification accuracy for MSI status is worse when applied to datasets with differing characteristics, which could be the case when applying these methods across different health systems, regions and populations. With continued experimentation, improvement, and validation of existing models, the use of machine learning to predict MSI may reach a level of accuracy sufficient for clinical application in the future.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Araghi, M.; Soerjomataram, I.; Jenkins, M.; Brierley, J.; Morris, E.; Bray, F.; Arnold, M. Global trends in colorectal cancer mortality: Projections to the year 2035. Int. J. Cancer 2019, 144, 2992–3000.
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164.
- Lal, N.; White, B.S.; Goussous, G.; Pickles, O.; Mason, M.J.; Beggs, A.D.; Taniere, P.; Willcox, B.E.; Guinney, J.; Middleton, G. KRAS Mutation and Consensus Molecular Subtypes 2 and 3 Are Independently Associated with Reduced Immune Infiltration and Reactivity in Colorectal Cancer. Clin. Cancer Res. 2018, 24, 224–233.
- Becht, E.; De Reyniès, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautès-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066.
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356.
- Seppälä, T.T.; Bohm, J.; Friman, M.; Lahtinen, L.; Väyrynen, V.M.J.; Liipo, T.K.E.; Ristimäki, A.P.; Kairaluoma, M.V.J.; Kellokumpu, I.H.; Kuopio, T.H.I.; et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br. J. Cancer 2015, 112, 1966–1975.
- Sjoquist, K.M.; Renfro, L.A.; Simes, R.J.; Tebbutt, N.C.; Clarke, S.; Seymour, M.T.; Adams, R.; Maughan, T.S.; Saltz, L.; Goldberg, R.M.; et al. Personalizing Survival Predictions in Advanced Colorectal Cancer: The ARCAD Nomogram Project. J. Natl. Cancer Inst. 2018, 110, 638–648.
- Hynes, S.O.; Coleman, H.G.; Kelly, P.J.; Irwin, S.; O’Neill, R.F.; Gray, R.T.; Mcgready, C.; Dunne, P.D.; McQuaid, S.; James, J.A.; et al. Back to the future: Routine morphological assessment of the tumour microenvironment is prognostic in stage II/III colon cancer in a large population-based study. Histopathology 2017, 71, 12–26.
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711.
- Rozek, L.S.; Schmit, S.L.; Greenson, J.K.; Tomsho, L.P.; Rennert, H.S.; Rennert, G.; Gruber, S.B. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival from Colorectal Cancer. J. Natl. Cancer Inst. 2016, 108.
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007, 18, 85–98.
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer—The stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162.
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531.
- Boland, P.M.; Yurgelun, M.B.; Boland, C.R. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 2018, 68, 217–231.
- Sinicrope, F.A.; Rego, R.L.; Halling, K.C.; Foster, N.; Sargent, D.J.; La Plant, B.; French, A.J.; Laurie, J.A.; Goldberg, R.M.; Thibodeau, S.N.; et al. Prognostic Impact of Microsatellite Instability and DNA Ploidy in Human Colon Carcinoma Patients. Gastroenterology 2006, 131, 729–737.
- Samowitz, W.S.; Curtin, K.; Ma, K.N.; Schaffer, D.; Coleman, L.W.; Leppert, M.; Slattery, M.L. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol. Biomark. Prev. 2001, 10, 917–923.
- Maoz, A.; Dennis, M.; Greenson, J.K. The Crohn’s-Like Lymphoid Reaction to Colorectal Cancer-Tertiary Lymphoid Structures with Immunologic and Potentially Therapeutic Relevance in Colorectal Cancer. Front. Immunol. 2019, 10, 1884.
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191.
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor Microsatellite-Instability Status as a Predictor of Benefit from Fluorouracil-Based Adjuvant Chemotherapy for Colon Cancer. N. Engl. J. Med. 2003, 349, 247–257.
- De Vos tot Nederveen Cappel, W.H.; Meulenbeld, H.J.; Kleibeuker, J.H.; Nagengast, F.M.; Menko, F.H.; Griffioen, G.; Cats, A.; Morreau, H.; Gelderblom, H.; Vasen, H.F.A. Survival after adjuvant 5-FU treatment for stage III colon cancer in hereditary nonpolyposis colorectal cancer. Int. J. Cancer 2004, 109, 468–471.
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective Mismatch Repair as a Predictive Marker for Lack of Efficacy of Fluorouracil-Based Adjuvant Therapy in Colon Cancer. J. Clin. Oncol. 2010, 28, 3219–3226.
- Webber, E.M.; Kauffman, T.L.; O’Connor, E.; Goddard, K.A. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer 2015, 15, 1–12.
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 2017, 19, 187–225.
- Shaikh, T.; Handorf, E.A.; Meyer, J.E.; Hall, M.J.; Esnaola, N.F. Mismatch Repair Deficiency Testing in Patients with Colorectal Cancer and Nonadherence to Testing Guidelines in Young Adults. JAMA Oncol. 2018, 4, e173580.
- Greenson, J.K.; Huang, S.-C.; Herron, C.; Moreno, V.; Bonner, J.D.; Tomsho, L.P.; Ben-Izhak, O.; Cohen, H.I.; Trougouboff, P.; Bejhar, J.; et al. Pathologic Predictors of Microsatellite Instability in Colorectal Cancer. Am. J. Surg. Pathol. 2009, 33, 126–133.
- Jenkins, M.A.; Hayashi, S.; O’Shea, A.-M.; Burgart, L.J.; Smyrk, T.C.; Shimizu, D.; Waring, P.M.; Ruszkiewicz, A.R.; Pollett, A.F.; Redston, M.; et al. Pathology Features in Bethesda Guidelines Predict Colorectal Cancer Microsatellite Instability: A Population-Based Study. Gastroenterology 2007, 133, 48–56.
- Greenson, J.K.; Bonner, J.D.; Ben-Yzhak, O.; Cohen, H.I.; Miselevich, I.; Resnick, M.B.; Trougouboff, P.; Tomsho, L.D.; Kim, E.; Low, M.; et al. Phenotype of Microsatellite Unstable Colorectal Carcinomas: Well-Differentiated and Focally Mucinous Tumors and the Absence of Dirty Necrosis Correlate With Microsatellite Instability. Am. J. Surg. Pathol. 2003, 27, 563–570.
- Walsh, M.D.; Cummings, M.C.; Buchanan, D.D.; Dambacher, W.M.; Arnold, S.; McKeone, D.; Byrnes, R.; Barker, M.A.; Leggett, B.A.; Gattas, M.; et al. Molecular, Pathologic, and Clinical Features of Early-Onset Endometrial Cancer: Identifying Presumptive Lynch Syndrome Patients. Clin. Cancer Res. 2008, 14, 1692–1700.
- Alexander, J.; Watanabe, T.; Wu, T.-T.; Rashid, A.; Li, S.; Hamilton, S.R. Histopathological Identification of Colon Cancer with Microsatellite Instability. Am. J. Pathol. 2001, 158, 527–535.
- Buckowitz, A.; Knaebel, H.-P.; Benner, A.; Bläker, H.; Gebert, J.; Kienle, P.; Doeberitz, M.V.K.; Kloor, M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br. J. Cancer 2005, 92, 1746–1753.
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520.
- Williams, D.S.; Bird, M.J.; Jorissen, R.N.; Yu, Y.L.; Walker, F.; Zhang, H.H.; Nice, E.C.; Burgess, A.W. Nonsense Mediated Decay Resistant Mutations Are a Source of Expressed Mutant Proteins in Colon Cancer Cell Lines with Microsatellite Instability. PLoS ONE 2010, 5, e16012.
- Willis, J.A.; Reyes-Uribe, L.; Chang, K.; Lipkin, S.M.; Vilar, E. Immune Activation in Mismatch Repair–Deficient Carcinogenesis: More Than Just Mutational Rate. Clin. Cancer Res. 2020, 26, 11–17.
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; De La Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J. Natl. Cancer Inst. 2004, 96, 261–268.
- Hyde, A.; Fontaine, D.; Stuckless, S.; Green, R.; Pollett, A.; Simms, M.; Sipahimalani, P.; Parfrey, P.; Younghusband, B. A Histology-Based Model for Predicting Microsatellite Instability in Colorectal Cancers. Am. J. Surg. Pathol. 2010, 34, 1820–1829.
- Fujiyoshi, K.; Yamaguchi, T.; Kakuta, M.; Takahashi, A.; Arai, Y.; Yamada, M.; Yamamoto, G.; Ohde, S.; Takao, M.; Horiguchi, S.-I.; et al. Predictive model for high-frequency microsatellite instability in colorectal cancer patients over 50 years of age. Cancer Med. 2017, 6, 1255–1263.
- Roman, R.; Verdu, M.; Calvo, M.; Vidal, A.; Sanjuan, X.; Jimeno, M.; Salas, A.; Autonell, J.; Trias, I.; González, M.; et al. Microsatellite instability of the colorectal carcinoma can be predicted in the conventional pathologic examination. A prospective multicentric study and the statistical analysis of 615 cases consolidate our previously proposed logistic regression model. Virchows Arch. 2010, 456, 533–541.
- Bessa, X.; Alenda, C.; Paya, A.; Álvarez, C.; Iglesias, M.; Seoane, A.; Dedeu, J.M.; Abulí, A.; Ilzarbe, L.; Navarro, G.; et al. Validation Microsatellite Path Score in a Population-Based Cohort of Patients with Colorectal Cancer. J. Clin. Oncol. 2011, 29, 3374–3380.
- Brazowski, E.; Rozen, P.; Pel, S.; Samuel, Z.; Solar, I.; Rosner, G. Can a gastrointestinal pathologist identify microsatellite instability in colorectal cancer with reproducibility and a high degree of specificity? Fam. Cancer 2012, 11, 249–257.
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715.
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81.
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056.
- Steu, S.; Baucamp, M.; Von Dach, G.; Bawohl, M.; Dettwiler, S.; Storz, M.; Moch, H.; Schraml, P. A procedure for tissue freezing and processing applicable to both intra-operative frozen section diagnosis and tissue banking in surgical pathology. Virchows Arch. 2008, 452, 305–312.
- Sprung, R.W.; Brock, J.W.C.; Tanksley, J.P.; Li, M.; Washington, M.K.; Slebos, R.J.C.; Liebler, D.C. Equivalence of Protein Inventories Obtained from Formalin-fixed Paraffin-embedded and Frozen Tissue in Multidimensional Liquid Chromatography-Tandem Mass Spectrometry Shotgun Proteomic Analysis. Mol. Cell. Proteom. 2009, 8, 1988–1998.
- Yeh, Y.-C.; Nitadori, J.-I.; Kadota, K.; Yoshizawa, A.; Rekhtman, N.; Moreira, A.L.; Sima, C.S.; Rusch, V.W.; Adusumilli, P.S.; Travis, W.D. Using frozen section to identify histological patterns in stage I lung adenocarcinoma of ≤3 cm: Accuracy and interobserver agreement. Histopathology 2015, 66, 922–938.
- Ratnavelu, N.D.; Brown, A.P.; Mallett, S.; Scholten, R.J.; Patel, A.; Founta, C.; Galaal, K.; Cross, P.; Naik, R. Intraoperative frozen section analysis for the diagnosis of early stage ovarian cancer in suspicious pelvic masses. Cochrane Database Syst. Rev. 2016, 2016, 010360.
- Mantel, H.T.; Westerkamp, A.C.; Sieders, E.; Peeters, P.M.J.G.; De Jong, K.P.; Boer, M.T.; de Kleine, R.H.; Gouw, A.S.H.; Porte, R.J. Intraoperative frozen section analysis of the proximal bile ducts in hilar cholangiocarcinoma is of limited value. Cancer Med. 2016, 5, 1373–1380.
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Rev. Cancer 2020, 1, 789–799.
- Cao, R.; Yang, F.; Ma, S.-C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.-H.; Wang, T.; Lu, W.-J.; Cai, W.-J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080–11091.
- Schmauch, B.; Romagnoni, A.; Pronier, E.; Saillard, C.; Maillé, P.; Calderaro, J.; Kamoun, A.; Sefta, M.; Toldo, S.; Zaslavskiy, M.; et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020, 11, 1–15.
- Zhang, R.; Osinski, B.L.; Taxter, T.J.; Perera, J.; Lau, D.J.; Khan, A.A. Adversarial deep learning for microsatellite instability prediction from histopathology slides. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018.
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting Endometrial Cancer Subtypes and Molecular Features from Histopathology Images Using Multi-resolution Deep Learning Models. bioRxiv 2020.
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J.; et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020, 159, 1406–1416.e11.
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed Cell Death 1 (PD-1) and Its Ligand (PD-L1) in Common Cancers and Their Correlation with Molecular Cancer Type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970.
- Wang, H.; Wang, X.; Xu, L.; Zhang, J.; Cao, H. Analysis of the transcriptomic features of microsatellite instability subtype colon cancer. BMC Cancer 2019, 19, 605.
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708.
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, MA, USA, 18–23 June 2018; pp. 6848–6856.
- Quellec, G.; Cazuguel, G.; Cochener, B.; Lamard, M. Multiple-Instance Learning for Medical Image and Video Analysis. IEEE Rev. Biomed. Eng. 2017, 10, 213–234.
- Wang, T.; Lu, W.; Yang, F.; Liu, L.; Dong, Z.; Tang, W.; Chang, J.; Huan, W.; Huang, K.; Yao, J. Microsatellite Instability Prediction of Uterine Corpus Endometrial Carcinoma Based on H&E Histology Whole-Slide Imaging. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1289–1292.
- Khan, A.A. Generalizable and Interpretable Deep Learning Framework for Predicting MSI from Histopathology Slide Images. U.S. Patent 20190347557, 14 November 2019.




