
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | María J. Blanco Prieto | + 2614 word(s) | 2614 | 2021-03-09 04:40:00 | | | |
| 2 | Dean Liu | Meta information modification | 2614 | 2021-03-25 11:12:30 | | |
Video Upload Options
Extracellular vesicles (EVs) are constituted by a group of heterogeneous membrane vesicles secreted by most cell types that play a crucial role in cell–cell communication.
1. Extracellular Vesicles
1.1. Classification
EVs comprise submicron particles heterogeneous in size, delimited by a lipid bilayer that cannot replicate. Traditionally, they have been classified according to their size and biogenesis, distinguishing: small particles or exosomes of endosomal origin with diameters ranging from 30 to 150 nm; ectosomes or microvesicles directly shed from the plasma membrane and polydisperse in size (100–1000 nm); and apoptotic bodies generated as a consequence of programmed cell death (1000–5000 nm) [1][2]. However, in recent years it has become apparent that the picture is more complex than expected. Assigning an EV to a particular biogenesis pathway still remains extraordinarily difficult given the overlap in size-distribution and protein-expression patterns among different EV types, especially when referring to exosomes and microvesicles, challenging the attempts to define a more precise nomenclature for EV classes [3]. Consequently, the latest recommendations of the ISEV encourage authors to define EV subtypes considering their physical characteristics—attending to: (a) size (small, medium/large) or density (low, medium, high) according to a defined range, (b) biochemical composition relaying on specific markers (e.g., CD63, CD81, annexin V, etc.), (c) isolation conditions (e.g., hypoxia, serum conditioning), and/or d) cellular origin (platelet, endothelial, cardiomyocytes, etc.)—rather than by the use of the traditional terms exosomes or microvesicles [1]. A summary of EVs’ physical and biochemical properties as well as parental cell conditions is provided in Table 1. In this review, the term EVs will refer to both exosomes and microvesicles.
Table 1. Requirements for extracellular vesicles’ separation and classification based on their physical and biochemical properties as well as donor cell conditions.
| Physical properties | Size [1] |
Small EVs | <100 nm | |
| Small/medium EVs | <200 nm | |||
| Medium/large EVs | >200 nm | |||
| Density (in sucrose) [4] |
Low | 1.13–1.19 g/mL | ||
| Medium | 1.16–1.28 g/mL | |||
| High | >1.28 g/mL | |||
| Biochemical composition | Surface antigens [1] |
Tetraspanins MHC class I Integrins Transferrin receptor LAMP1/2 Heparan sulfate |
Proteoglycans EMMPRIN ADAM10 GPI-anchored 5ʹnucleotidase CD73 Complement-binding proteins CD55 and CD59 Sonic hedgehog |
|
| Lipids [1][5] |
Phosphatidylserine Phosphatidylinositol Phosphatidylethanolamine Phosphatidylcholine |
Cholesterol Ceramide Diacylglycerol Glycosphingolipids |
||
| Internal cargo [1][6][7] |
Proteins | TSG101 ALIX VPS4A/B ARRDC1 Flotillins-1 and 2 |
Caveolins Annexins Heat shock proteins HSC70 and HSP84 Syntenin |
|
| Cardiac-related miRNAs | let-7 miR-16 miR-17-92 miR-19b miR-20a/b miR-21a miR-24 miR-26a miR-34 miR-93 miR-94a miR-107a miR-125b miR-126 |
miR-130a/b miR-132 miR-143 miR-145 miR-146a miR-181b miR-182 miR-208a miR-210 miR-214 miR-294 miR-302a miR-451 |
||
| Conditions at EVs harvest | Cell culture conditions [1] |
Normoxia Hypoxia Surface coating |
Treatment Grade of confluency Passage number |
|
| Donor status [1] |
Age Biological sex Circadian variation Body mass index |
Pathological/healthy condition Exercise level Diet Medication |
||
ADAM10: ADAM metallopeptidase domain 10; ALIX: ALG-2-interacting protein X; ARRDC1: arrestin domain-containing protein 1; EMMPRIN: extracellular matrix metalloproteinase inducer; EVs: extracellular vesicles; GPI: glycosylphosphatidylinositol; LAMP: lysosomal-associated membrane protein; MHC: major histocompatibility complex; miRNA: microRNA; TSG101: tumor susceptibility gene 101; VPS4A/B: Vacuolar Protein Sorting 4 Homolog A/B.
1.2. Biology
EVs are released into the extracellular space by most cell types and can be found in a wide range of body fluids as reservoirs of lipids, proteins, nucleic acids, and carbohydrates of their parental cells. Current knowledge supports the view that each cell type tunes EVs’ biogenesis, depending on its activation status. Moreover, their cargo is particular to the stimulus or biological condition triggering their formation and release, suggesting the existence of intracellular selective cargo-sorting mechanisms. Subsequently, EVs’ composition will directly affect their fate and function [2][8][9]. There is a consensus about two major EV biogenesis pathways, giving rise to the most widely studied subpopulations, exosomes and microvesicles. Exosomes, generated within the endosomal system, are intraluminal vesicles formed by the inward budding of the endosomal membrane during maturation of multivesicular endosomes (MVEs) and are released to the extracellular space by fusion of MVEs with the cell membrane [10]. As for microvesicles, they originate by outward budding of the plasma membrane [11]. Microvesicle biogenesis determines the expression of surface-specific antigens from the cell origin, as well as the externalization of phosphatidylserine on the outer membrane leaflet [12], although the latter is not a prerequisite in all microvesicles [13]. Despite the aforementioned differences in biogenesis, the overlap in size, density, or composition, together with the lack of appropriate technology, hampers the possibility of distinguishing EV subpopulations once released to the extracellular medium, favoring the use of the generic term EVs, instead of a more specific nomenclature [1].
1.3. Mechanism of Action
The lipid bilayer protects EVs’ content from degradation by nucleases and proteinases present in biofluids, enabling the transfer of proteins, lipids, or nucleic acids from parental cells to recipient cells [14]. As such, EVs contribute to normal homeostasis, but also to the progression of several pathologies, including CVDs [15][16]. The mechanisms by which EVs mediate intercellular communication are not completely understood but are supposed to involve specific interactions between proteins or lipids enriched at the EVs surface (e.g., tetraspanins, integrins, lectins, phosphatidylserine) and receptors at the plasma membrane of recipient cells (e.g., intercellular adhesion molecules (ICAMs), annexin V, galectin 5) [2][17][18][19][20]. After docking at the cell membrane, EVs can remain at the binding site eliciting functional responses in recipient cells by activating downstream molecular pathways, or by direct interaction with extracellular matrix components [21] (Figure 1). They can also be internalized by endocytosis or by fusion with the plasma membrane undergoing different fates. For instance, endocytosed EVs can reach the MVEs and be targeted for degradation by lysosomes, they can escape digestion by back fusion with the MVEs’ membrane, or they can be re-secreted to the extracellular space via the early endocytic recycling pathway [21][22][23][24][25]. Either by direct fusion with the plasma membrane or after escaping lysosomal degradation, EVs can release their content into the cytoplasm of recipient cells and regulate cellular processes [26] (Figure 1).
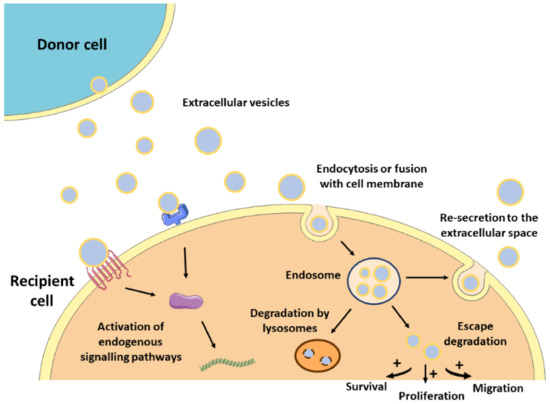
Figure 1. Mechanism of action of EVs. After released from donor cells, EVs may induce a response in recipient cells by different mechanisms. First, EVs may remain at the binding site on the cell membrane eliciting functional responses by activating downstream molecular pathways. Alternatively, EVs may be internalized by endocytosis or fusion with the cell membrane undergoing different intracellular fates. They can be targeted for degradation by lysosomes, they can escape degradation and modulate cell behavior, or they can be re-secreted to the extracellular space.
1.4. Separation and Characterization of Extracellular Vesicles
EVs can be separated from the cell culture medium and most body fluids (liquid biopsy), blood being the most frequently studied source of EVs. Before EV separation, some preanalytical parameters should be considered [27][28], such as the use of serum-free media or EV-depleted serum for EV separation from conditioned medium [1][29]. Moreover, differences in the physicochemical and biochemical properties of the selected separation methods can impact the enriched EV subpopulations [30][31][32][33][34] and do not enable an absolute purification of EVs from other contaminants [35].
Ultracentrifugation (UC) is the most commonly used EV separation and enrichment technique based on particle density, involving multiple centrifugation and ultracentrifugation steps [30]. Speeds of 10,000–20,000 g enable the separation of medium/large vesicles, while small-sized vesicles are recovered at higher speeds (100,000 g).
Size exclusion techniques include ultrafiltration and chromatography. Ultrafiltration is usually based on cellulose filters defined by molecular mass and size exclusion range [31], while size exclusion chromatography (SEC), separates fractions by elution with phosphate buffered saline (PBS) and has been proven to be reliable and scalable for various applications [36].
Immune affinity isolation is based on the immunolabeling of proteins on the surface of EVs, enabling the separation of specific particle subpopulations from other EV classes, contaminant protein aggregates or lipoproteins. Usually, specific antibodies are conjugated to magnetic beads and EVs are separated using magnets [37][38].
A range of commercial kits are also available, some based on polymer precipitation- methods [31][32][33] and others on non-precipitation alternatives, for instance, those selective for phosphatidylserine positive vesicles [29]. Alternative or complementary techniques to classical procedures are also emerging, including microfluidics, asymmetric flow field-flow fractionation, or high-resolution flow cytometry [35].
After separation, EVs’ purity should be tested by the use of multiple complementary methods: (i) western blotting to analyze EVs markers (e.g., CD63, Alix, etc.) and co-isolated contaminants [1]; (ii) nanoparticle tracking analysis (NTA), which can determine particle size and concentration [39]; (iii) conventional transmission electron microscopy (TEM) and the more strongly recommended cryo-TEM [40], or (iv) nanoflow cytometry, that enables the determination of cell surface antigens, the quantification of EV subpopulations based on parental cell markers [41][42], and the lipid nature of the studied particles with cell-permeant, non-fluorescent pro-dyes . Also, current advances in EV-adapted proteomic, lipidomic, and genomic technologies will greatly help to delimit the molecular signature of the EV subpopulations under research.
2. Potential Applications of Extracellular Vesicles as Therapeutic Agents in Myocardial Infarction
Driven by the drawbacks associated with cell transplantation as well as the key role of stem cell paracrine secretion in cardiac repair, EVs have emerged recently as a next-generation cell-free regenerative therapy. Several studies have been performed in the last five years, aiming to test EVs’ potential as cell substitutes in the cardiac regenerative field, with significant preclinical success (Figure 2). All these studies collect different cell sources, isolation techniques, therapeutic doses, or administration routes, reflecting the heterogeneity and immature nature of the field. Here, we group the most relevant findings from these studies as well as a brief compendium of the EV-associated molecules involved in heart repair, based on the parental cell type. A summary of these can be found in Table 2.
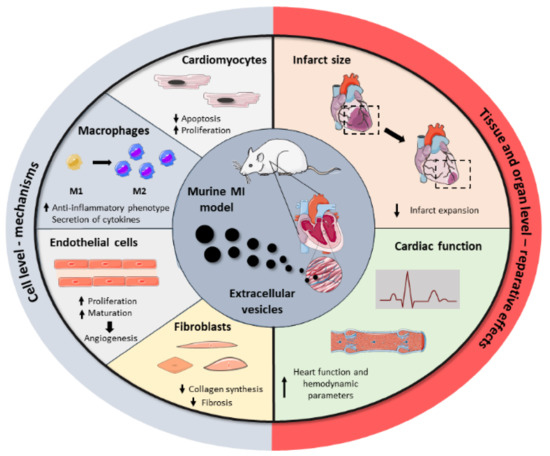
Figure 2. Summary of the beneficial effects of EVs in cardiac repair. Administration of EVs in MI preclinical models showed that EVs modulate a regenerative response in several cardiac cells, including cardiomyocytes, macrophages, endothelial cells, and fibroblasts. Together, these cell-level effects result in the reduction of infarct size and the improvement of cardiac function after MI.
Table 2. Representative preclinical efficacy studies from the last five years, using extracellular vesicles as therapeutic agents for myocardial infarction.
| Cell Source | Isolation Method | Animal Model | Dose | Administration Route and Time Post-MI | Reparative Effect | Molecule/Mechanism Involved | Ref |
|---|---|---|---|---|---|---|---|
| MSCs | |||||||
| Rat BM-MSCs | Total Exosome Isolation Kit (Invitrogen) | Rat, permanent | 20 µg | IM; immediate |
|
- | |
| Mouse BM-MSCs | Density- gradient UC | Mouse, I/R | 50 µg | IM; immediate after reperfusion |
|
Inhibition of TLR4 by miR-182 | |
| Proinflammatory rat BM-MSCs | Density-gradient UC | Mouse, permanent | 50 µg | IM; immediate |
|
Suppression of NF-κB and regulation of AKT1/AKT2 | |
| BM-MSCs | UC | Rat, permanent | 10 µg EVs (and 2×106 BM-MSCs) | IM; at 30 min |
|
- | |
| ATV-pre-treated rat BM-MSCs | UC | Rat, permanent | 10 µg | IM; immediate |
|
lncRNA H19 and miR-675 | |
| Mouse BM-MSCs | UC | Mouse, permanent | - | IV; immediate and day 6 |
|
miR-210 and Efna3 gene suppression | |
| Mouse BM-MSCs | UC | Mouse, permanent | EVs derived from 2×107 cells | IM; immediate |
|
miR-210 | |
| Rat BM-MSCs | Total Exosome Isolation Kit (Invitrogen) | Rat, I/R | 5 µg | IM; prior to reperfusion |
|
AMPK and AKT pathways | |
| Mouse BM-MSCs | UC | Mouse, I/R | 12.5 µg/ 5.62×105 EVs | IM; 24h prior to ischemia | • Decreased infarct size | Reduced expression of pro-apoptotic genes PDCD4, PTEN, Peli1 and FasL via miR-21a-5p | |
| Mouse BM-MSCs | UC | Mouse, permanent | 200 µg | IM; immediate |
|
miR-125b | |
| BM-MSCs | ExoQuick | Rat, permanent | - | IM; immediate |
|
miR-24 | |
| Rat ADSCs | UC | Rat, permanent | 2.5×1012 particles | IV; at 1h |
|
S1P/SK1/S1PR1 activation | |
| Rat ADSCs | Ultrafiltration and UC | Rat, I/R | 400 µg | IV; at reperfusion |
|
Wnt/β-catenin activation | |
| Human umbilical cord MSCs | Density-gradient UC | Rat, permanent | 400 µg and 800 µg | IV; once daily for 7 days | • Safety: no effect on hemolysis, no vascular and muscle stimulation, no side effects on hematology indexes, liver and renal function, and protective effect on weight loss | - | |
| Human umbilical cord MSCs | ExoQuick-TC (System Biosciences) | Rat, permanent | 400 µg | IM; immediate |
|
- | |
| Human umbilical cord MSCs | Density-gradient UC | Rat, permanent | 400 µg | IV; immediate |
|
Upregulation of Smad7 by inhibition of miR-125b-5p | |
| Cardiac MSCs | Precipitacion with PEG | Mouse, permanent | 50 µg | IM; immediate |
|
- | |
| CDCs | |||||||
| Human CDCs | Ultrafiltration and precipitation with PEG | Pig, I/R | 7.5 mg | IC; 30 min after reperfusion IM; 30 min after reperfusion |
|
- | |
| Porcine CDCs | Ultrafiltration followed by Field-Flow Fractionation | Pig, I/R | 9.16 mg | IM; at 72h after reperfusion |
|
- | |
| Human CDCs | Ultrafiltration and PEG precipitation | Pig, I/R | 7.5 mg | IM; at 20 min after reperfusion |
|
Regulation of gene expression by miRNA | |
| Human CDCs | Ultrafiltration and precipitation with PEG | Rat, I/R | 350 µg | IM; at 30 min after reperfusion |
|
- | |
| Human CDCs | ExoQuick (precipitation) | Rat, permanent | 250 µg | IM; at 4 weeks |
|
Regulation of gene expression by miRNA | |
| CPCs | |||||||
| Human CPCs | Density-gradient UC | Mice, permanent | 8 µg | IM; at 15 min |
|
Activation of endoglin in endothelial cells | |
| Rat CPCs | UC | Rat, I/R | 5 µg/kg | IM; during reperfusion |
|
Decreased levels of collagen I, collagen III, vimentin and CTGF Regulation of gene expression via miRNA |
|
| Human CPCs | UC | Rat, permanent and I/R | 1011 particles | IM; at 1h after permanent ligation or at reperfusion |
|
miR-146a-3p, miR-132, and miR-181a PAPP-A IGF-1 |
|
| iPS | |||||||
| Human iPS | UC | Mouse, permanent | 3×1010 particles | Transcutaneous echo-guided IM; at 3 weeks | • Increased cardiac function | Regulation of gene expression via miRNA | |
| Human iPS | UC | Mouse, permanent | 100 µg (1010 particles) | IM; at 2 days or 3 weeks |
|
- | |
| Mouse iPS | UC | Mouse, I/R | 100 µg | IM; at 48h after reperfusion |
|
Regulation of gene expression via miRNA and metabolic regulation via protein delivery (in silico analysis) |
|
| ESC | |||||||
| Human ESC | UC | Mouse, permanent | 20 µg | IM; immediate |
|
Targeting miR-497 by lncRNA MALAT1 | |
| Human ESC | UC | Mouse, permanent | - | Transcutaneous echo-guided IM; at 2-3 weeks |
|
Gene regulation of DNA repair, cell survival, cell cycle progression and cardiomyocyte contractility (in silico) | |
| Mouse ESC | UC | Mouse, permanent | - | IM; immediate |
|
Regulation of CPC cell cycle and association with proliferation and survival mediated by miR-294 | |
ADSCs: adipose tissue-derived mesenchymal stem cells; ATV: atorvastatin; BM-MSCs: bone marrow-derived mesenchymal stromal cells; CDCs: cardiosphere-derived cells; CPCs: cardiac progenitor cells; CTGF: connective tissue growth factor; ESC: embryonic stem cells; EVs: extracellular vesicles; I/R: ischemia/reperfusion; IC: intracoronary; IGF-1: insulin-like growth factor-1; IM: intramyocardially; iPS: induced pluripotent stem cells; IV: intravenously; lncRNA: long non-coding RNA; LV: left ventricle; MALAT1: metastasis-associated lung adenocarcinoma transcript 1; MI: myocardial infarction; miRNA: microRNA; MSCs: mesenchymal stromal cells; NF-κB: nuclear factor kappa B; PAPP-A: pregnancy-associated plasma protein A; PEG: polyethylene glycol; S1P: sphingosine 1-phosphate; S1PR1: sphingosine-1-phosphate receptor 1; SK1: sphingosine kinase 1; Smad7: mothers against decapentaplegic homolog 7; TLR4: toll-like receptor 4; UC: ultracentrifugation.
References
- World Health Organization Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 4 December 2020).
- Andreadou, I.; Cabrera-Fuentes, H.A.; Devaux, Y.; Frangogiannis, N.G.; Frantz, S.; Guzik, T.; Liehn, E.; Pedrosa da Costa Gomes, C.; Schulz, R.; Hausenloy, D.J. Immune cells as targets for cardioprotection: New players and novel therapeutic opportunities. Res. 2019, 115, 1117–1130.
- Abbafati, C.; Machado, D.B.; Cislaghi, B.; Salman, O.M.; Karanikolos, M.; McKee, M.; Abbas, K.M.; Brady, O.J.; Larson, H.J.; Trias-Llimós, S.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222, doi:10.1016/S0140-6736(20)30925-9.
- Burden, G.; Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Banach, M.; Barengo, N.C.; Beaton, A.Z.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. Am. Coll. Cardiol. 2020, 76, 2982–3021, doi:10.1016/j.jacc.2020.11.010.
- de Abreu, R.C.; Fernandes, H.; da Costa Martins, P.A.; Sahoo, S.; Emanueli, C.; Ferreira, L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Rev. Cardiol. 2020, 17, 685–697, doi:10.1038/s41569-020-0389-5.
- Marbán, E. A mechanistic roadmap for the clinical application of cardiac cell therapies. Biomed. Eng. 2018, 2, 353–361, doi:10.1038/s41551-018-0216-z.A.
- Marbán, E. The Secret Life of Exosomes: What Bees Can Teach Us About Next-Generation Therapeutics. Am. Coll. Cardiol. 2018, 71, 193–200, doi:10.1016/j.jacc.2017.11.013.
- Grigorian Shamagian, L.; Madonna, R.; Taylor, D.; Climent, A.M.; Prosper, F.; Bras-Rosario, L.; Bayes-Genis, A.; Ferdinandy, P.; Fernández-Avilés, F.; Izpisua Belmonte, J.C.; et al. Perspectives on Directions and Priorities for Future Preclinical Studies in Regenerative Medicine. Res. 2019, 124, 938–951, doi:10.1161/CIRCRESAHA.118.313795.
- Menasché, P. Cell therapy trials for heart regeneration—Lessons learned and future directions. Rev. Cardiol. 2018, 15, 659–671.
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Extracell. Vesicles 2018, 7, 1535750, doi:10.1080/20013078.2018.1535750.
- Whittaker, T.E.; Nagelkerke, A.; Nele, V.; Kauscher, U.; Stevens, M.M. Experimental artefacts can lead to misattribution of bioactivity from soluble mesenchymal stem cell paracrine factors to extracellular vesicles. Extracell. Vesicles 2020, 9, 1807674, doi:10.1080/20013078.2020.1807674.
- Sluijter, J.P.G.; Davidson, S.M.; Boulanger, C.M.; Buzás, E.I.; De Kleijn, D.P.V.; Engel, F.B.; Giricz, Z.; Hausenloy, D.J.; Kishore, R.; Lecour, S.; et al. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: Position Paper from the Working Group on Cellular Biology of the Heart of the European Society of Cardiology. Res. 2018, 114, 19–34, doi:10.1093/cvr/cvx211.
- Chong, S.Y.; Lee, C.K.; Huang, C.; Ou, Y.H.; Charles, C.J.; Richards, A.M.; Neupane, Y.R.; Pavon, M.V.; Zharkova, O.; Pastorin, G.; et al. Extracellular vesicles in cardiovascular diseases: Alternative biomarker sources, therapeutic agents, and drug delivery carriers. J. Mol. Sci. 2019, 20, 3272, doi:10.3390/ijms20133272.
- Martins-Marques, T.; Hausenloy, D.J.; Sluijter, J.P.G.; Leybaert, L.; Girao, H. Intercellular Communication in the Heart: Therapeutic Opportunities for Cardiac Ischemia. Trends Mol. Med. 2020, S1471-4914, 30263–X, doi:10.1016/j.molmed.2020.10.002.
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232, doi:10.1016/j.cell.2016.01.043.
- Ghafarian, F.; Pashirzad, M.; Khazaei, M.; Rezayi, M.; Hassanian, S.M.; Ferns, G.A.; Avan, A. The clinical impact of exosomes in cardiovascular disorders: From basic science to clinical application. Cell. Physiol. 2019, 234, 12226–12236, doi:10.1002/jcp.27964.
- Saludas, L.; Pascual-Gil, S.; Roli, F.; Garbayo, E.; Blanco-Prieto, M.J. Heart tissue repair and cardioprotection using drug delivery systems. Maturitas 2018, 110, 1–9, doi:10.1016/J.MATURITAS.2018.01.011.
- Rezaie, J.; Rahbarghazi, R.; Pezeshki, M.; Mazhar, M.; Yekani, F.; Khaksar, M.; Shokrollahi, E.; Amini, H.; Hashemzadeh, S.; Sokullu, S.E.; et al. Cardioprotective role of extracellular vesicles: A highlight on exosome beneficial effects in cardiovascular diseases. Cell. Physiol. 2019, 234, 21732–21745, doi:10.1002/jcp.28894.
- Tikhomirov, R.; Donnell, B.R.O.; Catapano, F.; Faggian, G.; Gorelik, J.; Martelli, F.; Emanueli, C. Exosomes: From Potential Culprits to New Therapeutic Promise in the Setting of Cardiac Fibrosis. Cells 2020, 9, 592, doi:10.3390/cells9030592.
- Deb, A.; Gupta, S.; Mazumder, P.B. Exosomes: A new horizon in modern medicine. Life Sci. 2020, 118623, doi:10.1016/j.lfs.2020.118623.
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Russell, A.E.; Sneider, A.; Witwer, K.W.; Bergese, P.; Bhattacharyya, S.N.; Cocks, A.; Cocucci, E.; Erdbrügger, U.; Falcon-Perez, J.M.; Freeman, D.W.; et al. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. Extracell. Vesicles 2019, 8, 1684862, doi:10.1080/20013078.2019.1684862.
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res. Int. 2018, 2018, doi:10.1155/2018/8545347.
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Drug Deliv. Rev. 2020, 159, 308–321, doi:10.1016/j.addr.2020.03.002.
- Femminò, S.; Penna, C.; Margarita, S.; Comità, S.; Brizzi, M.F.; Pagliaro, P. Extracellular vesicles and cardiovascular system: Biomarkers and Cardioprotective Effectors. Pharmacol. 2020, 135, 106790, doi:10.1016/j.vph.2020.106790.
- Kesidou, D.; da Costa Martins, P.A.; de Windt, L.J.; Brittan, M.; Beqqali, A.; Baker, A.H. Extracellular Vesicle miRNAs in the Promotion of Cardiac Neovascularisation. Physiol. 2020, 11, doi:10.3389/fphys.2020.579892.
- Pinheiro, A.; Silva, A.M.; Teixeira, J.H.; Gonçalves, R.M.; Almeida, M.I.; Barbosa, M.A.; Santos, S.G. Extracellular vesicles: Intelligent delivery strategies for therapeutic applications. Control. Release 2018, 289, 56–69.
- De Jong, B.; Barros, E.R.; Hoenderop, J.G.J.; Rigalli, J.P. Recent advances in extracellular vesicles as drug delivery systems and their potential in precision medicine. Pharmaceutics 2020, 12, 1006.
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Rev. Cell Dev. Biol. 2014, 30, 255–289.
- Taylor, J.; Azimi, I.; Monteith, G.; Bebawy, M. Ca2+ mediates extracellular vesicle biogenesis through alternate pathways in malignancy. Extracell. Vesicles 2020, 9, doi:10.1080/20013078.2020.1734326.
- Boulanger, C.M.; Loyer, X.; Rautou, P.E.; Amabile, N. Extracellular vesicles in coronary artery disease. Rev. Cardiol. 2017, 14, 259–272.
- Connor, D.E.; Exner, T.; Ma, D.D.F.; Joseph, J.E. The majority of circulating platelet-derived microparticles fail to bind annexin V, lack phospholipid-dependent procoagulant activity and demonstrate greater expression of glycoprotein Ib. Haemost. 2010, 103, 1044–1052, doi:10.1160/TH09-09-0644.
- Momen-Heravi, F.; Getting, S.J.; Moschos, S.A. Extracellular vesicles and their nucleic acids for biomarker discovery. Ther. 2018, 192, 170–187, doi:10.1016/j.pharmthera.2018.08.002.
- Cesselli, D.; Parisse, P.; Aleksova, A.; Veneziano, C.; Cervellin, C.; Zanello, A.; Beltrami, A.P. Extracellular vesicles: How drug and pathology interfere with their biogenesis and function. Physiol. 2018, 9, 1394.
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266, doi:10.1182/blood-2004-03-0824.
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678, doi:10.1158/0008-5472.CAN-09-2470.
- Christianson, H.C.; Svensson, K.J.; Van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Natl. Acad. Sci. USA 2013, 110, 17380–17385, doi:10.1073/pnas.1304266110.
- Fuentes, P.; Sesé, M.; Guijarro, P.J.; Emperador, M.; Sánchez-Redondo, S.; Peinado, H.; Hümmer, S.; Ramón y. Cajal, S. ITGB3-mediated uptake of small extracellular vesicles facilitates intercellular communication in breast cancer cells. Commun. 2020, 11, doi:10.1038/s41467-020-18081-9.
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of exosomes through receptor-mediated endocytosis. Cancer Res. 2019, 17, 337–347.
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687, doi:10.1111/j.1600-0854.2010.01041.x.
- Ares, G.R.; Ortiz, P.A. Dynamin2, clathrin,andlipid rafts mediate endocytosis of the apical Na/K/2Cl cotransporter NKCC2 in thick ascending limbs. Biol. Chem. 2012, 287, 37824–37834, doi:10.1074/jbc.M112.386425.
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes Derived from Epstein-Barr Virus-Infected Cells Are Internalized via Caveola-Dependent Endocytosis and Promote Phenotypic Modulation in Target Cells. Virol. 2013, 87, 10334–10347, doi:10.1128/jvi.01310-13.




