
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Simona Delle Monache | + 1318 word(s) | 1318 | 2021-03-03 10:08:36 | | | |
| 2 | Peter Tang | Meta information modification | 1318 | 2021-03-10 03:47:24 | | |
Video Upload Options
Liquid biopsy has emerged as a minimally invasive tool that is capable of detecting genomic alterations from primary or metastatic tumors, allowing the prognostic stratification of patients, the detection of the minimal residual disease after surgical or systemic treatments, the monitoring of therapeutic response, and the development of resistance, establishing an opportunity for early intervention before imaging detection or worsening of clinical symptoms. On the other hand, preclinical and clinical evidence demonstrated the role of gut microbiota dysbiosis in promoting inflammatory responses and cancer initiation. Altered gut microbiota is associated with resistance to chemo drugs and immune checkpoint inhibitors, whereas the use of microbe-targeted therapies including antibiotics, pre-probiotics, and fecal microbiota transplantation can restore response to anticancer drugs, promote immune response, and therefore support current treatment strategies in colorectal cancer (CRC).
1. Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related death and morbidity worldwide according to the global cancer statistics (GLOBOCAN) presented in 2018. The 5-year survival rate ranges from 90% to 14% if CRC is diagnosed at a localized or metastatic stage, respectively, and approximately 25% of CRC patients present metastatic disease at diagnosis, while almost half of them will develop metastases [1].
If early diagnosis and treatment of CRC can significantly improve the cure rate, traditional biomarkers (Carcino Embryonic Antigen (CEA), Carbohydrate Antigen 19-9 (CA19-9), Fecal Occult Blood Test (FOBT)) as well as colon/sigmoidoscopy do not fully satisfy clinical needs in CRC screening due to their lack in sensitivity and specificity [2]. Furthermore, primary tumor resection is eventually associated to adjuvant chemotherapy with fluoropyrimidines with or without oxaliplatin according to TNM stage and pathological risk factors in early CRC [3], does not always seem sufficient to eliminate circulating tumor cells (CTCs) and other components involved in establishing pre-metastatic niche-promoting immune evasion and maintenance of stemness [4].
Circulating tumor DNA (ctDNA) and RNAs and non-coding RNAs (ncRNAs) released into the bloodstream via microvescicles or tumor cell lysis represent, together with CTCs, different sides of the same coin: liquid biopsy. Liquid biopsy has emerged as a promising minimally invasive tool for precision medicine due to its ability to provide multiple global snapshots of primary and metastatic tumors at different times and more representative images of the spatial and temporal tumor heterogeneity [5] compared to tissue biopsy. In fact, even though tissue biopsy remains the gold standard for the histopatological definition and the molecular stratification of tumors, it is often difficult to perform, especially in relapsed and metastatic settings, and it does not support intratumoral heterogeneity and clonal evolutions related to driver mutations, which may occur during tumor development or treatment.
Among other elements potentially involved in cancer initiation, development, recurrence, and metastasis, one that only recently received its due attention is the host microbiota—and for CRC, especially the gut microbiota. The host microbiota is composed of bacteria (≈99%), viruses, and mycetes, existing in a condition of eubosis with the human body conferring important benefits related to physical and mental health, and the development of the individual [6]. In turn, this dynamic balance is affected by host genetics, lifestyle [7], and dietary habits [8] and gut microbiota dysbiosis may play a role in promoting inflammatory responses and alterations of the immunosurveillance, which can led to cancer initiation and/or progression [9] (Figure 1).
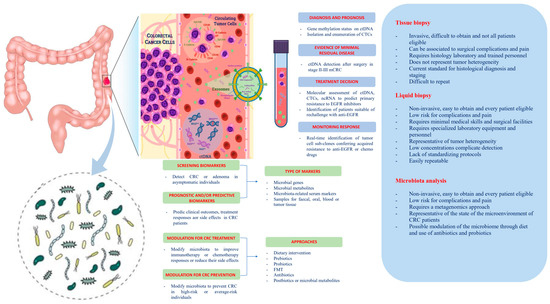
Figure 1. Potential clinical applications related to liquid biopsy and gut microbiota in colorectal cancer. Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), non-coding RNA (ncRNA), and exosomes are promising liquid biopsy markers for colorectal cancer with multiple potential advantages compared to tissue biopsy. CTCs from colorectal cancer (CRC) can be shed from the primary tumor into the bloodstream, which also contains ctDNA released from tumor tissue through apoptosis, necrosis, and secretion, as well as circulating normal DNA released from healthy tissue. NcRNAs (miRNAs and lncRNAs) encapsulated by exosomes can be actively secreted into the extracellular fluid by various types of cells in the tumor or passively released due to the apoptosis and necrosis of tumor cells and can eventually be found in the circulation. Besides liquid biopsy, several potential clinical applications for harnessing the gut microbiota in CRC include development of screening, prognostic and predictive biomarkers, and microbiota modulation for CRC prevention and treatment. FMT, fecal microbiota transplantation.
2. Liquid Biopsy
The term liquid biopsy refers to procedures of isolation of cancer-derived components such as CTCs, exosomes, ctDNA, ncRNAs, and proteins from peripheral blood or other body fluids, and their genomic or proteomic evaluation [10]. Assessment of such elements via non-invasive and low-risk blood-based detection tests could improve CRC screening, diagnosis, staging, and predict relapse and metastasis [11][12] and be effective in monitoring residual disease and drug resistance in CRC patients receiving systemic treatment [13][14].
2.1. Circulating Tumor Cells (CTCs)
CTCs are tumor cells released into the bloodstream from the primary tumor or metastases [15], which could escape from immune recognition and drug treatment, and subsequently form a niche in other tissues, promoting tumor recurrence and metastasis [16].
2.2. Circulating Tumor DNA (ctDNA)
Circulating tumor DNA (CtDNA) is a kind of double-stranded DNA, a fragment of cell-free DNA (cfDNA), that originates from active, apoptotic, necrotic, or circulating tumor cells. CtDNA retains epigenetic characteristics and harbors tumor-specific mutations detectable in the bloodstream and other body fluids [10][17]. Importantly, ctDNA half-life varies from several minutes to a few hours, and as for CTCs, its plasma levels depend on tumor load, ranging from 50% to 90% in non-metastatic and metastatic cancer patients, respectively [4][10][18][19]. Furthermore, healthy people and cancer patients can be distinguished according to the fragment length distribution pattern of cfDNA [17]. These data suggest that ctDNA analysis may represent a real-time tumor burden assessment.
2.3. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs)
MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are ncRNAs molecules involved in the regulation of protein-coding gene expression through mRNA degradation and silencing or activating and repressing genes via a variety of mechanisms at both transcriptional and translational levels. Both classes of ncRNAs regulate multiple cellular processes such as growth, development, and differentiation showing to be crucial for cancer initiation, progression, and dissemination and can be found in serum or other body fluids bound to protein or lipid complexes, or more frequently inside extracellular vescicles (i.e., exosomes) [20]. Furthermore, these elements seem to be strongly associated with the development of drug resistance in CRC [21][22][23][24]. For these reasons, miRNA and lncRNAs could have potential application in diagnosis, prognosis, and treatment of CRC.
3. Microbiota
The study of microbiota started several years ago, and multiple definitions have been conceived to explain its meaning [25]. In general, the terms “microbiota” and “microbiome” refer to the complex of organisms found within a specific environment and their genomic pool, respectively [26][27]. Thus, the human gut microbiota consists of a multitude of microorganisms colonizing the gut and existing in that complex state of dynamic equilibrium (i.e., eubiosis), which is made of reciprocal interactions and multiple networks between themselves and the host cells. This is an equilibrium with specific spatial and temporal characteristics, whose deregulation might lead to dysbiosis [26].
The human gut microbiota—with its thousands of different bacterial taxa, eucaryotic microbes, and virus together with the intestinal barrier—is a very selective and important filter for the well-being of the whole organism, and as a neuroendocrine structure today considered as a “second brain”, it is a component of the complex gut ecosystem [28][29]. The gastrointestinal microbiota varies according to the anatomical location and among individuals [11], and it plays different roles, from the supply of nutrients to the control of inflammation and carcinogenesis [26]. Commensal bacteria instruct the immune and physiological systems throughout life and are responsible for the presence of inflammatory and immune cells in the healthy intestine: the so-called “physiological” or “controlled” inflammation [30]. For this purpose, numerous evidence has demonstrated that a direct relationship between modification in the gut microbiota composition and some pathologies exist [31][32]. Among these diseases, obesity and metabolic alterations induced by some nutrients and diet, or autoimmune diseases such as type 1 diabetes and inflammatory bowel disease, are characterized by changes in the microbiome and gut dysbiosis [33].
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953.
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 845–856.
- van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. 2011, 728, 23–34.
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765.
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920.
- Gomez, A.; Espinoza, J.L.; Harkins, D.M.; Leong, P.; Saffery, R.; Bockmann, M.; Torralba, M.; Kuelbs, C.; Kodukula, R.; Inman, J.; et al. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host Microbe 2017, 22, 269–278.e263.
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89.
- Graf, D.; Di Cagno, R.; Fak, F.; Flint, H.J.; Nyman, M.; Saarela, M.; Watzl, B. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Health Dis. 2015, 26, 26164.
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704.
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484.
- Lampis, A.; Hahne, J.C.; Hedayat, S.; Valeri, N. MicroRNAs as mediators of drug resistance mechanisms. Curr. Opin. Pharmacol. 2020, 54, 44–50.
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Science 2019, 110, 1148–1155.
- Kilgour, E.; Rothwell, D.G.; Brady, G.; Dive, C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020, 37, 485–495.
- Masuda, T.; Hayashi, N.; Iguchi, T.; Ito, S.; Eguchi, H.; Mimori, K. Clinical and biological significance of circulating tumor cells in cancer. Mol. Oncol. 2016, 10, 408–417.
- Romiti, A.; Raffa, S.; Di Rocco, R.; Roberto, M.; Milano, A.; Zullo, A.; Leone, L.; Ranieri, D.; Mazzetta, F.; Medda, E.; et al. Circulating tumor cells count predicts survival in colorectal cancer patients. J. Gastrointest. Liver Dis. JGLD 2014, 23, 279–284.
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flugen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. As. Med. 2019, 69, 107–122.
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.O.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389.
- Musella, V.; Pietrantonio, F.; Di Buduo, E.; Iacovelli, R.; Martinetti, A.; Sottotetti, E.; Bossi, I.; Maggi, C.; Di Bartolomeo, M.; de Braud, F.; et al. Circulating tumor cells as a longitudinal biomarker in patients with advanced chemorefractory, RAS-BRAF wild-type colorectal cancer receiving cetuximab or panitumumab. Int. J. Cancer 2015, 137, 1467–1474.
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990.
- Tang, X.J.; Wang, W.; Hann, S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019, 163, 58–72.
- Qi, F.F.; Yang, Y.; Zhang, H.; Chen, H. Long non-coding RNAs: Key regulators in oxaliplatin resistance of colorectal cancer. Biomed. Pharmacother. 2020, 128, 110329.
- Wei, L.; Wang, X.; Lv, L.; Liu, J.; Xing, H.; Song, Y.; Xie, M.; Lei, T.; Zhang, N.; Yang, M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer 2019, 18, 147.
- Wang, X.; Jiang, G.; Ren, W.; Wang, B.; Yang, C.; Li, M. LncRNA NEAT1 Regulates 5-Fu Sensitivity, Apoptosis and Invasion in Colorectal Cancer Through the MiR-150-5p/CPSF4 Axis. OncoTargets Ther. 2020, 13, 6373–6383.
- Xian, Z.; Hu, B.; Wang, T.; Zeng, J.; Cai, J.; Zou, Q.; Zhu, P. lncRNA UCA1 Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells Through miR-23b-3p/ZNF281 Axis. OncoTargets Ther. 2020, 13, 7571–7583.
- Brussow, H. Problems with the concept of gut microbiota dysbiosis. Microb. Biotechnol. 2020, 13, 423–434.
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S38–S44.
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31.
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214.
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65.
- O’Keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706.
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964.
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86.
- Gagniere, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518.




