
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Asahiro Morishita | + 2891 word(s) | 2891 | 2021-02-25 05:05:47 | | | |
| 2 | Peter Tang | Meta information modification | 2891 | 2021-03-07 09:15:24 | | |
Video Upload Options
Hepatocellular carcinoma (HCC) is the seventh most frequent cancer and the fourth leading cause of cancer mortality worldwide. MicroRNAs (miRNAs) are small, endogenous, noncoding, single-stranded RNAs that modulate the expression of their target genes at the posttranscriptional and translational levels. Aberrant expression of miRNAs has frequently been detected in cancer-associated genomic regions or fragile sites in various human cancers and has been observed in both HCC cells and tissues. The precise patterns of aberrant miRNA expression differ depending on disease etiology, including various causes of hepatocarcinogenesis, such as viral hepatitis, alcoholic liver disease, or nonalcoholic steatohepatitis.
1. Introduction
Liver cancer is the sixth most frequent malignancy and the fourth leading cause of cancer-related deaths worldwide [1]. Hepatocellular carcinoma (HCC) accounts for approximately 80% of all liver cancers and is a main cause of cancer mortality [2]. The risk factors of hepatocarcinogenesis include various liver diseases, such as infections caused by hepatitis B virus (HBV) and hepatitis C virus (HCV), alcoholic liver disease (ALD), nonalcoholic steatohepatitis (NASH), and nonalcoholic fatty liver disease (NAFLD) [3][4][5]. Despite recent great progress in HCC therapy, the 5-year survival rate for advanced-stage HCC remains poor owing to its late diagnosis, resistance to anticancer therapy, and high frequency of recurrence [6][7][8]. Therefore, elucidating the detailed underlying mechanisms and pathogenesis of HCC is important for the development of new diagnostic and prognostic biomarkers and therapeutic drugs.
MicroRNAs (miRNAs, miRs) are small, endogenous, interfering, noncoding RNAs of 21–30 nucleotides in length. More than 2600 miRNAs have been predicted to be encoded by the human genome, with the ability to modulate more than 15,000 genes [9]. Each miRNA negatively regulates target genes by binding to the 3′ untranslated region (UTR) of mRNAs. These complexes are involved in RNA-mediated interference, and in vertebrates, mRNA transcripts are usually not cleaved by a miRNA-associated RNA-induced silencing complex (RISC) but rather undergo translational repression and degradation via deadenylation [10]. The miRNA-associated RISC can suppress gene expression [11][12]. Remarkably, a single miRNA can modulate more than 200 mRNAs [13][14].
The biogenesis of miRNAs includes several steps, such as transcription, cleavage, export, further cleavage, strand selection, and interaction with mRNAs [15] (Figure 1). Generally, RNA polymerase II transcribes primary miRNA (pri-miRNAs) transcripts in the canonical pathway or mirtron pathway [16]. The 5′ ends of pri-miRNAs are capped, and the 3′ ends are polyadenylated [17]. Pri-miRNAs are then cleaved to 70–100-nucleotide hairpin-structured precursors (pre-miRNAs) by a nuclear RNase III enzyme named Drosha, which is a double-stranded RNA-binding domain protein involving DiGeorge syndrome critical region 8 (DGCR8)/Pasha as a cofactor [18]. Next, pre-miRNAs are exported from the nucleus to the cytoplasm, after binding with exportin-5 and Ran-GTP [19] and are cleaved by Dicer [20], with transactivation response element RNA-binding 70 protein (TRBP) serving as a cofactor for Dicer [20]. Finally, double-stranded pre-miRNAs undergo rapid unwinding, when loaded onto Argonaute (AGO), and only one strand, which serves as a guide to target mRNAs, remains bound [21]. These RISCs with the AGO protein modulate mRNA degradation and translational inhibition [21].
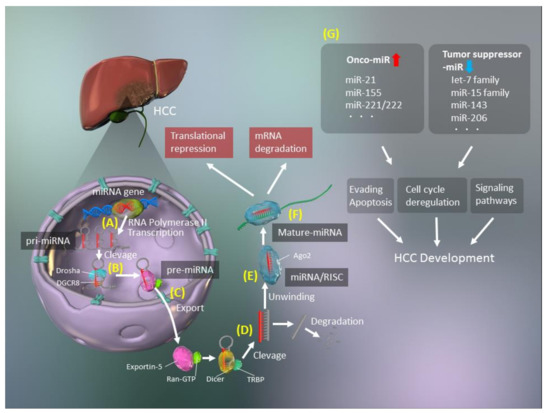
Figure 1. Schematic of microRNA (miRNA, miR) biogenesis. (A) Synthesis of primary miRNA (pri-miRNA) transcripts from genomic DNA by RNA polymerase II. (B) The pri-miRNA is cleaved by Drosha/DiGeorge syndrome critical region 8 (DGCR8) and processed to a precursor miRNA (pre-miRNA). (C) The pre-miRNA forms a complex with exportin-5 and Ran-GTP and is exported to the cytoplasm. (D) The exported hairpin pre-miRNA is cleaved by Dicer/transactivation response element RNA-binding 70 protein (TRBP). (E) The double-stranded miRNA is unwound and forms an RNA-induced silencing complex (RISC) with Argonaute 2 (AGO2). (F) The miRNA is separated into a mature, single-stranded miRNA. (G) Upregulation of oncogenic miRs (oncomiRs) and downregulation of tumor suppressor miRs promote hepatocellular carcinoma (HCC) development.
However, the biogenesis of miRNAs is more diverse and may involve noncanonical pathways that bypass Drosha/DGCR8 processing. There are several noncanonical types of miRNAs, including mirtrons, small nucleolar RNAs (snoRNAs), transfer RNAs (tRNAs), and short hairpin RNAs (shRNAs) [22]. Mirtrons are pre-miRNAs that are generated without Drosha by pre-mRNA splicing and intron debranching [23][24]. SnoRNAs [25] and tRNAs [26][27] have different internal hairpin structures, and their processing involves the cleavage activity of Dicer, without Drosha/DGCR8. Transcripts of shRNA, which are derived from unannotated and intergenic regions, can function as Dicer substrates during transcription [28][29].
Although the important roles of miRNAs in the modulation of mRNA expression are well established, their precise functions remain elusive. Interestingly, miRNAs modulate the expression of approximately 30% of all human genes, many of which are tumor-associated or are in regions of instability in the genome [30][31]. There is clear evidence of key roles for miRNAs in human carcinogenesis [13][32][33][34][35], with two types of miRNAs identified, namely, oncogenic miRNAs (oncomiRs) and tumor suppressor miRNAs (Figure 1). OncomiRs induce carcinogenesis by inhibiting the expression of tumor suppressors, while tumor suppressor miRNAs inhibit oncogene expression in normal cells and are lacking in cancer cells. Two miRNAs, miR-15 and miR-16, were first reported to be altered in cancer and are associated with a frequently targeted chromosomal deletion of BCL2, which encodes an anti-apoptotic factor [36].
Several reports have demonstrated relationships between miRNAs and HCC [35][37][38][39][40] and identified potential miRNA biomarkers and therapeutic targets for HCC diagnosis and treatment.
2. Roles of miRNAs in the Liver
2.1. Lipid Metabolism
The liver plays a critical role in lipid metabolism. Dysfunctions in lipid metabolism induce excessive accumulation of hepatic triglycerides and fatty acids, resulting in various liver diseases, such as NAFLD and NASH. Several miRNAs have pivotal functions in the maintenance of cholesterol and fatty acid metabolism [41], and others, such as miR-33, miR-103, miR-104, and miR-307, act as modulators of lipid and cholesterol levels [42].
Various serum miRNAs, including miR-122, miR-21, miR-34a, and miR-451, are enhanced in patients with NAFLD [43]. A representative liver-specific miRNA, miR-122, which is highly upregulated in the liver, is involved in hepatic cholesterol and lipid metabolism in this disease [44]. Suppression of miR-122 can diminish the plasma cholesterol levels, reduce hepatic fatty acid and cholesterol synthesis, and increase the oxidation of hepatic fatty acids [45]. In addition, miR-122 is associated with hepatic lipogenesis-related enzymes, including fatty acid synthase (FASN) and acetyl-CoA carboxylase (ACC1). The inhibition of miR-122 also decreases hepatic lipogenesis in obese mice [44].
Several reports have demonstrated that miR-34a is associated with hepatic steatosis [46][47]. This miRNA inhibits hepatic silent information regulator 1 (sirtuin 1, SIRT1), peroxisome proliferator-activated receptor-α (PPARα), and liver X receptor (LXR) [46][47][48]. Reduced SIRT1 levels in the liver of patients with NAFLD can be recovered by the inhibition of miR-34a, which results in the improvement of hepatic steatosis via PPARα and the activation of AMP-activated protein kinase (AMPK) [47].
2.2. Glucose Metabolism
miR-375, a novel islet-specific miRNA, can inhibit glucose-induced insulin secretion, whereas inhibition of miR-375 can increase insulin secretion. This miR-375-associated insulin modulation is independent of altered glucose metabolism and is related to insulin exocytosis. Myotrophin (MTPN), which is a target of miR-375, is also involved in glucose metabolism. The inhibition of MTPN enhances glucose-induced insulin secretion and insulin exocytosis, indicating that miR-375 might be a new therapeutic target for diabetes mellitus and NAFLD [49]. Expression of miR-23a is increased in a NASH-related HCC murine model, and overexpression of miR-23a via the interleukin-6 (IL-6)/STAT3 signaling pathway decreases glucose production through the inhibition of PGC1α and G6PC expression [50]. In addition, miR-143, which is involved in insulin resistance, controls the ORP8-dependent regulatory pathway of AKT, and miR-143 overexpression reduces insulin-stimulated AKT activation [51]. Moreover, miR-206 decreases lipid and glucose levels in hepatocytes by modulating lipogenesis and insulin signaling [52], which suggests that miR-206 might be a diagnostic biomarker and therapeutic target for NAFLD and hyperglycemia.
2.3. Hepatic Inflammation
Hepatic inflammation, which involves inflammatory cytokine production and endoplasmic reticulum stress, results from an abnormal immune response; the latter is mediated by several factors, such as viral and bacterial infections, metabolic disorders, alcohol abuse, drug allergies, and toxic reagents [53]. Various miRNAs, such as miR-122 and miR-132, play pivotal roles in the innate and adaptive immunity involved in hepatic inflammation. miR-122 is the most representative miRNA in the liver, and its loss results in inflammation, fibrosis, and HCC, indicating that miR-122 modulates anti-inflammatory effects [54][55]. Furthermore, miR-122 may inhibit hepatic infiltration of inflammatory cells and the secretion of various cytokines, including IL-6 and tumor necrosis factor-α (TNFα), by these cells [54][55]. miR-132, which inhibits the expression of SIRT1, is a mediator of inflammation in chronic liver diseases. The overexpression of miR-132 induces the translocation of nuclear factor-κB (NF-κB) into the nucleus, acetylation of p65, and production of IL-8 and MCP-1. The loss of miR-132, which is mediated by serum deprivation, diminishes the acetylation level of p65 and partially downregulates the expression of IL-8 and MCP-1. Therefore, the inhibition of miR-132 has anti-inflammatory effects in the liver.
2.4. Hepatic Fibrosis
Fibrosis is developed in the liver as a result of continuous and severe hepatocyte damage, resulting in an inflammatory cytokine storm. Several miRNAs can synergistically modulate inflammatory signaling pathways. Furthermore, several miRNAs are associated with the activation of hepatic stellate cells (HSCs) and progression of hepatic fibrosis via the regulation of related signaling pathways.
Members of the miR-29 family induce apoptosis via the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway and modulate the accumulation of the extracellular matrix (ECM) [56][57][58]. Stimulation of HSCs by transforming growth factor-beta (TGF-β) promotes myofibroblastic transition and ECM induction, resulting in liver fibrogenesis. TGF-β1 mediates the downregulation of miR-29 in HSCs [57]. In addition, miR-29 overexpression in mouse HSCs leads to the reduction of collagen-1α1 and collagen-4α1 levels [57][59][60] via modulation of various ECM genes. Similar to other hepatic fibrosis-related miRNAs, members of the miR-34 family promote hepatic fibrosis by activating HSCs, whereas those of the miR-378 family inhibit the development of fibrosis in a GLIS-dependent manner. The miR-15 family is associated with the induction of cell proliferation and apoptosis, and its members regulate the TGF-β signaling pathway by inhibiting TGFβR1, SMAD3, SMAD7, p38, and endoglin in cardiac fibrosis [61]. The miR-199 and miR-200 families are involved in the secretion of profibrotic cytokines and are responsible for ECM deposition [62][63]. These miRNA families act as modulators of hepatic fibrosis by targeting genes involved in fibrosis-related signaling pathways and activating HSCs.
3. Roles of miRNAs in Various Liver Diseases Leading to HCC
3.1. HBV Infection
HBV infection is one of the most prevalent risk factors of HCC development [64]. Thus, HBV-related HCC is a serious health concern worldwide [64]. Recently, many reports have demonstrated that miRNAs play pivotal roles in each stage of HBV-related HCC development [65][66][67][68]. HBV dysregulates miRNAs that modulate the expression of the host/HBV genes during HCC pathogenesis. In fact, this type of HCC is characterized by a range of immune response failures due to the dysregulation of miRNAs [69].
The HBV X protein (HBx) inhibits miR-34 expression via p53 stimulation in hepatocytes, which results in the upregulation of a macrophage-derived chemokine (CCL22), stimulation of regulatory T cells (Tregs), and the suppression of effector T cells, thereby increasing HBV genome transcription [70][71]. HBx-induced upregulation of miR-155 leads to a reduction in the suppressor of cytokine signaling-1 (SOCS1) expression, increasing JAK/STAT signaling and suppressing HBV infection mediated by the induction of interferon (IFN) signaling [72]. In addition, HBx-induced miR-155 blocks CCAAT/enhancer-binding protein (C/EBP), which activates the HBV enhancer (Enh) 11/core promoter and then inhibits HBV replication [73].
Liver-specific miR-122 is upregulated in the serum after HBV infection and is regarded as one of the key modulators of HBV replication [74][75][76]. However, Wang et al. [77] demonstrated that HBx suppressed miR-122 expression and increased the levels of HBV transcripts by blocking p53 binding to the HBV Enh1/core promoter. Another report has revealed that miR-122 blocks HBV pregenome RNA, which encodes the hepatitis B core antigen and viral polymerase and inhibits HBV replication via dysregulation of heme oxygenase-1 (HO-1), which suppresses refill of HBV covalently closed circular DNA [78]. In addition, upregulation of several miRNAs, including miR-184, miR-185, miR-196a, miR-199a-3p, miR-210, and miR-217, directly affects HBV transcription [79]. Furthermore, it has recently been demonstrated that the HBV virion produces its own HBV-miR-2 and HBV-miR-3 [80]. HBV-miR-3 inhibits mRNA expression of the hepatitis B core (HBc) protein, which is involved in HBV self-regulation [81], to prolong survival by escaping the host immune system [79][82]. miR-372 also modulates HBV expression, which depends on target pathways. HBx-upregulated miR-372 [83][84] targets the cyclic AMP response element-binding protein (CREB) by binding to HBV Enh1/core promoter (ENI-Cp), thereby suppressing HBV transcription. miR-372 targets nuclear factor 1 B-type protein (NFIB), which modulates HBV Enh1/ENI-Cp, thus increasing HBV transcript levels [79][83]. Various miRNAs are thus related to the modulation of HBV transcript levels and the host immune response to the virus.
3.2. HCV Infection
HCV contributes to serious liver problems, such as liver cancer, including HCC [85]. IFN-free, direct-acting antiviral therapy improves HCV treatment, with sustained virologic response rates greater than 95%, without many side effects [86]. However, the high mutation rate of HCV may result in the resistance to direct-acting antivirals, and patients with mutated HCV have low sustained virologic response rates [87]. Therefore, further studies of the molecular mechanism underlying HCV infections are needed to improve HCV therapy.
Dysregulation of miRNAs due to HCV infection occurs via multiple pathways, such as the immune response, lipid metabolism, and cell-cycle pathways [88]. Several target genes, such as PPARG and fibronectin 1 (FN1), were found to be downregulated, while other target genes, including stearoyl-CoA desaturase (SCD) and CREB1, were found to be upregulated by at least 11 miRNAs, namely, miR-130a/b, miR-200, miR-34a, miR-23b, miR-24, miR-146a, miR-381, miR-25, miR-200a, and miR-371-5p, after HCV infection [89]. In addition, miR-122 modulates the host immune response by regulating the expression of target genes and directly targeting the HCV genome [90][91][92]. In particular, miR-122 stabilizes the 5′ and 3′ UTRs of the HCV genome, and inhibition of this miRNA dramatically reduces the replication of HCV RNA [93][94]. By contrast, miR-141, miR-192, miR-215, and miR-491 increase HCV replication. Thus, miR-141 enhances HCV replication by reducing the tumor suppressor deleted in liver cancer 1 (DLC-1) [95], and miR-491 enables HCV entry via the PI3K/AKT signaling pathway [96]. However, miR-196, miR-29, let-7b, miR-130a, and miR-27a induce anti-HCV activity [59][97][98][99][100][101]. It has also been found that miR-199a suppresses HCV replication by blocking domain II of the internal ribosomal entry site [97]. In addition, IFN-β enhances the expression of several miRNAs with anti-HCV activity, such as miR-196, miR-431, and miR-448. Among these, miR-196 and miR-448 may directly target the HCV RNA genome [91]. These findings suggest that several miRNAs are strongly involved in the modulation of HCV infection and replication.
3.3. ALD
ALD is a complex disease caused by prolonged and heavy alcohol consumption, along with predisposing genetic factors. ALD can cause liver dysfunction, including steatohepatitis, fibrosis, cirrhosis, and eventually HCC [102]. There is tremendous evidence of the effects of miRNA dysregulation on the pathogenesis and development of ALD, such as liver damage, lipid metabolism dysfunction, inflammation, oxidative stress, apoptosis, and fibrosis [102]. Inflammation-related miRNAs, such as miR-132, miR-155, miR-146, and miR-21, influence alcohol/lipopolysaccharide (LPS)/TLR4 pathways. TLR4 transmits proinflammatory stimuli via a mitogen-activated protein kinases (MAPKs) or TIR domain-containing adaptor-inducing IFN-β (TRIF) [103]. The expression of miR-212 is increased by alcohol in intestinal cells, and LPS is also induced by alcohol in the liver via the suppression of ZO-1 [104][105]. Alcohol-induced oxidative stress downregulates miR-199a in liver sinusoidal endothelial cells by upregulating ET1 and hypoxia-inducible factor 1α (HIF1α), which are related to steatohepatitis and fibrosis in the liver [106]. The downregulation of miR-223 increases neutrophil infiltration in the liver and induces liver injury by inhibiting the IL-6/p47phox/reactive oxygen species axis [107]. The enhanced expression of miR-217 exacerbates alcoholic fatty changes by damaging SIRT1 and lipin-1 [108]. Moreover, upregulation of miR-34a in ALD results in the development of liver fibrosis via caspase-2, SIRT1, and matrix metallopeptidase (MMP) 1 and MMP2 [109]. In addition, miR-122 modulates hepatic lipid metabolism and inflammation [110]. The miRNA let-7 is diminished in response to alcohol and loss of let-7 induces a mesenchymal phenotype in HSCs to enhance liver injury by inhibiting LIN28B. Finally, an imbalance between let-7 and LIN28/28B causes oncogenesis [111].
3.4. NAFLD and NASH
NAFLD is defined as steatosis in at least 5% of hepatocytes [112], without other liver diseases, including viral hepatitis and autoimmune, alcohol-related, and genetic liver diseases. Recently, NAFLD has become one of the most frequent liver diseases worldwide [113]. Various miRNAs have been identified to be involved in NASH development. For example, the levels of miR-122, which is a representative hepatic miRNA, are 7.2 times higher in patients with NASH than in healthy subjects and 3.1 times higher than in patients with simple steatosis [114]. Liver-specific miR-122-knockout mice rapidly develop NASH and exhibit enhanced lipogenesis, changes in lipid secretion, IL-6 and TNF-α production, and upregulation of chemokine (C–C motif) ligand 2 (CCL2). Decreased miR-122 expression enhances fibrogenesis by inducing HIF1α and MAPK1, which can also facilitate HCC development [115]. In addition, miR-192 is involved in the development of TGF-β1-promoted fibrosis, which activates SMAD signaling [116]. However, members of a miRNA superfamily that includes miR-16, miR-497, miR-195, miR-322, and miR-15, particularly miR-15 and miR-16, regulate hepatic fibrosis and hepatocarcinogenesis [61]. The development and progression of NASH increases the risk of HCC via miRNAs. A recent report has demonstrated that miRNAs are pivotal for the activation of HSCs during NASH development [117]. Free cholesterol is accumulated because of increases in both SREBP2 and miR-33a signaling via the inhibition of PPARγ signaling, along with HSC activation and disruption of the SREBP2-induced cholesterol feedback system [117]. Upregulation of miR-21, which downregulates the expression of the tumor suppressor phosphatase and tensin homolog (PTEN), is mediated by unsaturated fatty acids in hepatocytes [118]. Furthermore, the expression of miR-155, which inhibits another tumor suppressor gene, C/EBPβ, is enhanced in mice fed a choline-deficient, amino acid-defined diet [73][119].
References
- Aly, A.; Ronnebaum, S.M.; Patel, D.; Doleh, Y.; Benavente, F. Epidemiologic, humanistic and economic burden of hepatocellular carcinoma in the USA: A systematic literature review. Hepatic Oncol. 2020, 7, HEP27.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549.
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604.
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64, 1577–1586.
- Altekruse, S.F.; Henley, S.J.; Cucinelli, J.E.; McGlynn, K.A. Changing Hepatocellular Carcinoma Incidence and Liver Cancer Mortality Rates in the United States. Am. J. Gastroenterol. 2014, 109, 542–553.
- Zhang, G.; Li, R.; Deng, Y.; Zhao, L. Conditional survival of patients with hepatocellular carcinoma: Results from the Surveillance, Epidemiology, and End Results registry. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 515–523.
- Xu, L.; Kim, Y.; Spolverato, G.; Gani, F.; Pawlik, T.M. Racial disparities in treatment and survival of patients with hepatocellular carcinoma in the United States. Hepatobiliary Surg. Nutr. 2016, 5, 43–52.
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302.
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. Epigenetic alteration and microRNA dysregulation in cancer. Front. Genet. 2013, 4, 258.
- Davis-Dusenbery, B.N.; Hata, A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes Cancer 2010, 1, 1100–1114.
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in Human Cancer. Adv. Exp. Med. Bio. 2013, 774, 1–20.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500.
- Wang, X.W.; Heegaard, N.H.H.; Orum, H. MicroRNAs in Liver Disease. Gastroenterology 2012, 142, 1431–1443.
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139.
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966.
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240.
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science 2009, 326, 1275–1279.
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744.
- Kim, Y.; Kim, V.N. MicroRNA Factory: RISC Assembly from Precursor MicroRNAs. Mol. Cell 2012, 46, 384–386.
- Xia, J.; Zhang, W. Noncanonical MicroRNAs and Endogenous siRNAs in Lytic Infection of Murine Gammaherpesvirus. PLoS ONE 2012, 7, e47863.
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell 2007, 130, 89–100.
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86.
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human snoRNA with MicroRNA-Like Functions. Mol. Cell 2008, 32, 519–528.
- Blelloch, R. Small RNAs—Their biogenesis, regulation and function in embryonic stem cells. StemBook 2008.
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785.
- Babiarz, J.E.; Hsu, R.; Melton, C.; Thomas, M.; Ullian, E.M.; Blelloch, R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 2011, 17, 1489–1501.
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009.
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587.
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 1–24.
- Michael, M.Z.; Connor, S.M.O.; Pellekaan, N.G.V.H.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003, 1, 882–891.
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 2007, 120, 1046–1054.
- Takahashi, T.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756.
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658.
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Nonlinear partial differential equations and applications: Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529.
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.-G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 Is a Target of miR-122a, a MicroRNA Frequently Down-regulated in Human Hepatocellular Carcinoma. Cancer Res. 2007, 67, 6092–6099.
- Wong, Q.W.; Lung, R.W.; Law, P.T.; Lai, P.B.; Chan, K.Y.; To, K.; Wong, N. MicroRNA-223 Is Commonly Repressed in Hepatocellular Carcinoma and Potentiates Expression of Stathmin1. Gastroenterology 2008, 135, 257–269.
- Varnholt, H.; Drebber, U.; Schulze, F.; Wedemeyer, I.; Schirmacher, P.; Dienes, H.-P.; Odenthal, M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008, 47, 1223–1232.
- Huang, X.-H.; Wang, Q.; Chen, J.-S.; Fu, X.-H.; Chen, X.-L.; Chen, L.-Z.; Li, W.; Bi, J.; Zhang, L.-J.; Fu, Q.; et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol. Res. 2009, 39, 786–794.
- Yu, G.; Yang, Z.; Peng, T.; Lv, Y. Circular RNAs: Rising stars in lipid metabolism and lipid disorders. J. Cell. Physiol. 2020.
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250.
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103.
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008, 48, 1810–1820.
- Clarke, J.; Sharapova, T.; Lake, A.D.; Blomme, E.; Maher, J.; Cherrington, N.J. Circulating microRNA 122 in the methionine and choline-deficient mouse model of non-alcoholic steatohepatitis. J. Appl. Toxicol. 2014, 34, 726–732.
- Salvoza, N.C.; Klinzing, D.C.; Gopez-Cervantes, J.; Baclig, M.O. Association of Circulating Serum miR-34a and miR-122 with Dyslipidemia among Patients with Non-Alcoholic Fatty Liver Disease. PLoS ONE 2016, 11, e0153497.
- Ding, J.; Li, M.; Wan, X.; Jin, X.; Chen, S.; Yu, C.; Li, Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 13729.
- Braza-Boïls, A.; Marí-Alexandre, J.; Molina, P.; Arnau, M.A.; Barceló-Molina, M.; Domingo, D.; Girbes, J.; Giner, J.; Martínez-Dolz, L.; Zorio, E. Deregulated hepatic microRNAs underlie the association between non-alcoholic fatty liver disease and coronary artery disease. Liver Int. 2016, 36, 1221–1229.
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230.
- Wang, B.; Hsu, S.-H.; Frankel, W.; Ghoshal, K.; Jacob, S.T. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating Glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology 2012, 56, 186–197.
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature 2011, 13, 434–446.
- Wu, H.; Zhang, T.; Pan, F.; Steer, C.J.; Li, Z.; Chen, X.; Song, G. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J. Hepatol. 2017, 66, 816–824.
- Liu, X.L.; Cao, H.X.; Fan, J.G. MicroRNAs as biomarkers and regulators of nonalcoholic fatty liver disease. J. Dig. Dis. 2016, 17, 708–715.
- Hsu, S.-H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; La Perle, K.; Chivukula, R.R.; et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883.
- Wen, J.; Friedman, J.R. miR-122 regulates hepatic lipid metabolism and tumor suppression. J. Clin. Investig. 2012, 122, 2773–2776.
- Mann, J.; Chu, D.C.; Maxwell, A.; Oakley, F.; Zhu, N.; Tsukamoto, H.; Mann, D.A. MeCP2 Controls an Epigenetic Pathway That Promotes Myofibroblast Transdifferentiation and Fibrosis. Gastroenterology 2010, 138, 705–714.e4.
- Roderburg, C.; Urban, G.-W.; Bettermann, K.; Vucur, M.; Zimmermann, H.W.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2010, 53, 209–218.
- Sekiya, Y.; Ogawa, T.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem. Biophys. Res. Commun. 2011, 412, 74–79.
- Bandyopadhyay, S.; Friedman, R.C.; Marquez, R.T.; Keck, K.; Kong, B.; Icardi, M.S.; Brown, K.E.; Burge, C.B.; Schmidt, W.N.; Wang, Y.; et al. Hepatitis C Virus Infection and Hepatic Stellate Cell Activation Downregulate miR-29: miR-29 Overexpression Reduces Hepatitis C Viral Abundance in Culture. J. Infect. Dis. 2011, 203, 1753–1762.
- Huang, J.; Yu, X.; Fries, J.W.U.; Zhang, L.; Odenthal, M. MicroRNA Function in the Profibrogenic Interplay upon Chronic Liver Disease. Int. J. Mol. Sci. 2014, 15, 9360–9371.
- Tijsen, A.J.; Van Der Made, I.; van den Hoogenhof, M.M.; Wijnen, W.J.; Van Deel, E.D.; De Groot, N.E.; Alekseev, S.; Fluiter, K.; Schroen, B.; Goumans, M.-J.; et al. The microRNA-15 family inhibits the TGFβ-pathway in the heart. Cardiovasc. Res. 2014, 104, 61–71.
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The Progression of Liver Fibrosis Is Related with Overexpression of the miR-199 and 200 Families. PLoS ONE 2011, 6, e16081.
- Sun, X.; He, Y.; Ma, T.-T.; Huang, C.; Zhang, L.; Li, J. Participation of miR-200a in TGF-β1-mediated hepatic stellate cell activation. Mol. Cell. Biochem. 2014, 388, 11–23.
- Mak, D.; Babb De Villiers, C.; Chasela, C.; Urban, M.I.; Kramvis, A. Analysis of risk factors associated with hepatocellular carcinoma in black South Africans: 2000–2012. PLoS ONE 2018, 13, e0196057.
- Jiang, J.; Gusev, Y.; Aderca, I.; Mettler, T.A.; Nagorney, D.M.; Brackett, D.J.; Roberts, L.R.; Schmittgen, T.D. Association of MicroRNA Expression in Hepatocellular Carcinomas with Hepatitis Infection, Cirrhosis, and Patient Survival. Clin. Cancer Res. 2008, 14, 419–427.
- Furuta, M.; Kozaki, K.-I.; Tanaka, S.; Arii, S.; Imoto, I.; Inazawa, J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2010, 31, 766–776.
- Hatziapostolou, M.; Polytarchou, C.; Aggelidou, E.; Drakaki, A.; Poultsides, G.A.; Jaeger, S.A.; Ogata, H.; Karin, M.; Struhl, K.; Hadzopoulou-Cladaras, M.; et al. An HNF4α-miRNA Inflammatory Feedback Circuit Regulates Hepatocellular Oncogenesis. Cell 2011, 147, 1233–1247.
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545.
- Sartorius, K.; Makarova, J.A.; Sartorius, B.; An, P.; Winkler, C.A.; Chuturgoon, A.A.; Kramvis, A. The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis. Cells 2019, 8, 1504.
- Xie, K.-L.; Zhang, Y.-G.; Liu, J.; Zeng, Y.; Wu, H. MicroRNAs Associated With HBV Infection And HBV-related HCC. Theranostics 2014, 4, 1176–1192.
- Yang, P.; Li, Q.-J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y.; et al. TGF-β-miR-34a-CCL22 Signaling-Induced Treg Cell Recruitment Promotes Venous Metastases of HBV-Positive Hepatocellular Carcinoma. Cancer Cell 2012, 22, 291–303.
- Su, C.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol. J. 2011, 8, 354.
- Wang, B.; Majumder, S.; Nuovo, G.; Kutay, H.; Volinia, S.; Patel, T.; Schmittgen, T.D.; Croce, C.; Ghoshal, K.; Jacob, S.T. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 2009, 50, 1152–1161.
- Arataki, K.; Hayes, C.N.; Akamatsu, S.; Akiyama, R.; Abe, H.; Tsuge, M.; Miki, D.; Ochi, H.; Hiraga, N.; Aikata, H.; et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med Virol. 2013, 85, 789–798.
- Nakamura, M.; Kanda, T.; Jiang, X.; Haga, Y.; Takahashi, K.; Wu, S.; Yasui, S.; Nakamoto, S.; Yokosuka, O. Serum microRNA-122 and Wisteria floribunda agglutinin-positive Mac-2 binding protein are useful tools for liquid biopsy of the patients with hepatitis B virus and advanced liver fibrosis. PLoS ONE 2017, 12, e0177302.
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457.
- Wang, S.; Qiu, L.; Yan, X.; Jin, W.; Wang, Y.; Chen, L.; Wu, E.; Ye, X.; Gao, G.F.; Wang, F.; et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G1-modulated P53 activity. Hepatolgy 2012, 55, 730–741.
- Qiu, L.; Fan, H.; Jin, W.; Zhao, B.; Wang, Y.; Ju, Y.; Chen, L.; Chen, Y.; Duan, Z.; Meng, S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 2010, 398, 771–777.
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552.
- Fisicaro, P.; Valdatta, C.; Boni, C.; Massari, M.; Mori, C.; Zerbini, A.; Orlandini, A.; Sacchelli, L.; Missale, G.; Ferrari, C. Early kinetics of innate and adaptive immune responses during hepatitis B virus infection. Gut 2009, 58, 974–982.
- Mizuguchi, Y.; Takizawa, T.; Yoshida, H.; Uchida, E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol. Res. 2016, 46, 391–406.
- Bertoletti, A.; Kennedy, P.T.F. The immune tolerant phase of chronic HBV infection: New perspectives on an old concept. Cell. Mol. Immunol. 2014, 12, 258–263.
- Bertoletti, A.; Ferrari, C. Adaptive immunity in HBV infection. J. Hepatol. 2016, 64, S71–S83.
- Busca, A.; Kumar, A. Innate immune responses in hepatitis B virus (HBV) infection. Virol. J. 2014, 11, 22.
- Torres, H.A.; Shigle, T.L.; Hammoudi, N.; Link, J.T.; Samaniego, F.; Kaseb, A.; Mallet, V. The oncologic burden of hepatitis C virus infection: A clinical perspective. CA A Cancer J. Clin. 2017, 67, 411–431.
- Bachofner, J.; Valli, P.V.; Bergamin, I.; Kröger, A.; Künzler, P.; Baserga, A.; Braun, D.L.; Seifert, B.; Moncsek, A.; Fehr, J.; et al. Excellent outcome of direct antiviral treatment for chronic hepatitis C in Switzerland. Swiss Med Wkly. 2018, 148, w14560.
- Shoukry, N.H. Hepatitis C Vaccines, Antibodies, and T Cells. Front. Immunol. 2018, 9, 1480.
- Ura, S.; Honda, M.; Yamashita, T.; Ueda, T.; Takatori, H.; Nishino, R.; Sunakozaka, H.; Sakai, Y.; Horimoto, K.; Kaneko, S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatolgy 2009, 49, 1098–1112.
- Steuerwald, N.M.; Parsons, J.C.; Bennett, K.; Bates, T.C.; Bonkovsky, H.L. Parallel microRNA and mRNA expression profiling of (genotype 1b) human hepatoma cells expressing hepatitis C virus. Liver Int. 2010, 30, 1490–1504.
- Kerr, T.A.; Korenblat, K.M.; Davidson, N.O. MicroRNAs and liver disease. Transl. Res. 2011, 157, 241–252.
- Pedersen, I.M.; Cheng, G.; Wieland, S.; Volinia, S.; Croce, C.M.; Chisari, F.V.; David, M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 2007, 449, 919–922.
- Peng, X.; Li, Y.; Walters, K.-A.; Rosenzweig, E.R.; Lederer, S.L.; Aicher, L.D.; Proll, S.; Katze, M.G. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genom. 2009, 10, 373.
- Lanford, R.E.; Hildebrandt-Eriksen, E.S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Orum, H. Therapeutic Silencing of MicroRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science 2009, 327, 198–201.
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science 2005, 309, 1577–1581.
- Banaudha, K.; Kaliszewski, M.; Korolnek, T.; Florea, L.; Yeung, M.L.; Jeang, K.-T.; Kumar, A. MicroRNA silencing of tumor suppressor DLC-1 promotes efficient hepatitis C virus replication in primary human hepatocytes. Hepatolgy 2011, 53, 53–61.
- Ishida, H.; Tatsumi, T.; Hosui, A.; Nawa, T.; Kodama, T.; Shimizu, S.; Hikita, H.; Hiramatsu, N.; Kanto, T.; Hayashi, N.; et al. Alterations in microRNA expression profile in HCV-infected hepatoma cells: Involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem. Biophys. Res. Commun. 2011, 412, 92–97.
- Murakami, Y.; Aly, H.H.; Tajima, A.; Inoue, I.; Shimotohno, K. Regulation of the hepatitis C virus genome replication by miR-199a. J. Hepatol. 2009, 50, 453–460.
- Hou, W.; Tian, Q.; Zheng, J.; Bonkovsky, H.L. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatolgy 2010, 51, 1494–1504.
- Cheng, J.-C.; Yeh, Y.-J.; Tseng, C.-P.; Hsu, S.-D.; Chang, Y.-L.; Sakamoto, N.; Huang, H.-D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. 2012, 69, 2621–2633.
- Chowdhury, J.B.; Shrivastava, S.; Steele, R.; Di Bisceglie, A.M.; Ray, R.; Ray, R.B. Hepatitis C Virus Infection Modulates Expression of Interferon Stimulatory Gene IFITM1 by Upregulating miR-130A. J. Virol. 2012, 86, 10221–10225.
- Shirasaki, T.; Honda, M.; Shimakami, T.; Horii, R.; Yamashita, T.; Sakai, Y.; Sakai, A.; Okada, H.; Watanabe, R.; Murakami, S.; et al. MicroRNA-27a Regulates Lipid Metabolism and Inhibits Hepatitis C Virus Replication in Human Hepatoma Cells. J. Virol. 2013, 87, 5270–5286.
- Li, H.-D.; Du, X.-S.; Huang, H.-M.; Chen, X.; Yang, Y.; Huang, C.; Meng, X.-M.; Li, J. Noncoding RNAs in alcoholic liver disease. J. Cell. Physiol. 2019, 234, 14709–14720.
- Mandrekar, P.; Szabo, G. Signalling pathways in alcohol-induced liver inflammation. J. Hepatol. 2009, 50, 1258–1266.
- Gao, B.; Bataller, R. Alcoholic Liver Disease: Pathogenesis and New Therapeutic Targets. Gastroenterology 2011, 141, 1572–1585.
- Tang, Y.; Banan, A.; Forsyth, C.B.; Fields, J.Z.; Lau, C.K.; Zhang, L.J.; Keshavarzian, A. Effect of Alcohol on miR-212 Expression in Intestinal Epithelial Cells and Its Potential Role in Alcoholic Liver Disease. Alcohol. Clin. Exp. Res. 2008, 32, 355–364.
- Yeligar, S.; Tsukamoto, H.; Kalra, V.K. Ethanol-Induced Expression of ET-1 and ET-BR in Liver Sinusoidal Endothelial Cells and Human Endothelial Cells Involves Hypoxia-Inducible Factor-1α and MicroRNA-199. J. Immunol. 2009, 183, 5232–5243.
- Li, M.; He, Y.; Zhou, Z.; Ramirez, T.; Gao, Y.; Gao, Y.; Ross, R.A.; Cao, H.; Cai, Y.; Xu, M.; et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6–p47phox–oxidative stress pathway in neutrophils. Gut 2017, 66, 705–715.
- Yin, H.; Liang, X.; Jogasuria, A.; Davidson, N.O.; You, M. miR-217 Regulates Ethanol-Induced Hepatic Inflammation by Disrupting Sirtuin 1–Lipin-1 Signaling. Am. J. Pathol. 2015, 185, 1286–1296.
- Wan, Y.; McDaniel, K.; Wu, N.; Ramos-Lorenzo, S.; Glaser, T.; Venter, J.; Francis, H.; Kennedy, L.; Sato, K.; Zhou, T.; et al. Regulation of Cellular Senescence by miR-34a in Alcoholic Liver Injury. Am. J. Pathol. 2017, 187, 2788–2798.
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98.
- McDaniel, K.; Huang, L.; Sato, K.; Wu, N.; Annable, T.; Zhou, T.; Ramos-Lorenzo, S.; Wan, Y.; Huang, Q.; Francis, H.; et al. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J. Biol. Chem. 2017, 292, 11336–11347.
- Nuño-Lámbarri, N.; Domínguez-Pérez, M.; Baulies-Domenech, A.; Monte, M.J.; Marin, J.J.G.; Rosales-Cruz, P.; Souza, V.; Miranda, R.U.; Bucio, L.; Montalvo-Jave, E.E.; et al. Liver Cholesterol Overload Aggravates Obstructive Cholestasis by Inducing Oxidative Stress and Premature Death in Mice. Oxidative Med. Cell. Longev. 2016, 2016, 9895176.
- Bernal-Reyes, R.; Castro-Narro, G.; Malé-Velázquez, R.; Carmona-Sánchez, R.; González-Huezo, M.; García-Juárez, I.; Chávez-Tapia, N.; Aguilar-Salinas, C.; Aiza-Haddad, I.; Ballesteros-Amozurrutia, M.; et al. Consenso mexicano de la enfermedad por hígado graso no alcohólico. Rev. Gastroenterol. México 2019, 84, 69–99.
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.M.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S.C. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 8001–8002.
- Wu, F.-L.; Jin, W.-B.; Li, J.-H.; Guo, A.-G. Targets for human encoded microRNAs in HBV genes. Virus Genes 2011, 42, 157–161.
- Coll, M.; El Taghdouini, A.; Perea, L.; Mannaerts, I.; Vila-Casadesús, M.; Blaya, D.; Rodrigo-Torres, D.; Affò, S.; Morales-Ibanez, O.; Graupera, I.; et al. Integrative miRNA and Gene Expression Profiling Analysis of Human Quiescent Hepatic Stellate Cells. Sci. Rep. 2015, 5, 11549.
- Tomita, K.; Teratani, T.; Suzuki, T.; Shimizu, M.; Sato, H.; Narimatsu, K.; Okada, Y.; Kurihara, C.; Irie, R.; Yokoyama, H.; et al. Free cholesterol accumulation in hepatic stellate cells: Mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 2014, 59, 154–169.
- Vinciguerra, M.; Sgroi, A.; Veyrat-Durebex, C.; Rubbia-Brandt, L.; Buhler, L.H.; Foti, M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology 2009, 49, 1176–1184.
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Boldin, M.P.; Taganov, K.D.; Nicoll, J.; Paquette, R.L.; Baltimore, D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008, 205, 585–594.




