
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivana Škrlec | + 1695 word(s) | 1695 | 2021-02-25 04:19:52 | | | |
| 2 | Peter Tang | Meta information modification | 1695 | 2021-03-06 16:38:25 | | |
Video Upload Options
The hepatitis E virus (HEV) is a positive single-stranded, icosahedral, quasi-enveloped RNA virus in the genus Orthohepevirus of the family Hepeviridae. Orthohepevirus A is the most numerous species of the genus Orthohepevirus and consists of eight different HEV genotypes that can cause infection in humans. HEV is a pathogen transmitted via the fecal–oral route, most commonly by consuming fecally contaminated water. A particular danger is the HEV-1 genotype, which poses a very high risk of vertical transmission from the mother to the fetus.
1. Introduction
Hepatitis E virus (HEV) is a positive single-stranded, small, quasi-enveloped RNA virus. It is the only member of the genus Orthohepevirus of the family Hepeviridae [1][2]. Orthohepevirus A is the largest species of the genus Orthohepevirus and consists of eight different genotypes of HEV that can cause infection in humans (HEV-1, 2, 3, 4, and 7), rabbits (HEV-3), pigs (HEV-3 and 4), boars (HEV-3, 4, 5, and 6), deer (HEV-3), mongoose (HEV-3), camel (HEV-7 and HEV-8), and yak (HEV-4) [3][4], as shown in Table 1.
Table 1. Classification of hepatitis E virus (HEV).
|
Genus |
Orthohepevirus |
|
Family |
Hepeviridae |
|
Species |
A, B, C, D |
|
Genotypes |
1,2,3,4,5,6,7,8 |
|
Virion |
32–34 nm, icosahedral |
|
Envelope |
quasi-enveloped |
|
Genome |
single-stranded, positive-sense, linear RNA |
|
Genome size |
7.2 kb |
|
Sensitivity |
Inactivation at temperatures above 70 °C |
In northern India, during the 1978 hepatitis epidemic, the hepatitis E virus was first identified in the Kashmir Valley. In that specific outbreak there were 5200 cases of hepatitis, resulting in 17,000 deaths [5].
The structure of the HEV genome was thoroughly studied in 1991. The RNA molecule consists of 7200 positively directed nucleotides [6]. There are three main coding regions whose protein products have not been thoroughly studied. It was also found that there is a fourth coding region characteristic of the HEV-1 genotype [6][7][8]. The heparan sulfate proteoglycan, HSC70 (heat shock cognate 70 kDa), allows HEV to enter the cell. Other specific, but insufficiently studied, receptors and co-receptors are also crucial for entering the cell. The process of releasing, transcribing, and translating HEV RNA, as well as the assemblage of new virions, occurs in the cytoplasm [6][9][10].
2. Molecular Biology of the Hepatitis E Virus
The hepatitis E virus genome is a single-stranded positive linear RNA 7.2 kb in length [11][12][13]. HEV RNA consists of four open reading frames (ORF) and untranslated regions at the 5’ and 3’ ends. At the 5’ end is a 7-methylguanosine cap and at the 3’ end is a polyadenylated tail, just like eukaryotic messenger RNA (mRNA) [14][15], as shown in Figure 1. During viral replication two viral RNA are generated: a full-length RNA of 7.2 kb and subgenomic RNA of 2.2 kb [13][14].
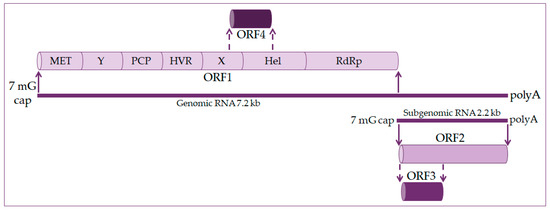
Figure 1. Hepatitis E virus genome organization. The genome is 7.2 kb positive-strand RNA with a 5′ 7-methylguanosine cap (7 mG cap) and a 3′ polyadenylated tail (polyA). It harbors four open reading frames (ORF). ORF1 codes nonstructural proteins, including a methyltransferase (MET), an RNA helicase (Hel), and an RNA-dependent RNA polymerase (RdRp), as well as less well-characterized domains, such as the Y domain, a papain-like cysteine protease (PCP), a hypervariable region (HVR), and the X domain. ORF2 and ORF3 encode the viral capsid and a small protein involved in virus secretion, respectively, translated from a 2.2 kb subgenomic RNA generated during viral replication. The ORF4 is found in the HEV-1 genotype and codes protein enhanced viral replication.
ORF1 occupies about 2/3 of the genome and codes nonstructural proteins included in viral RNA replication. These are methyltransferase (MET), X domain, helicase (Hel), and RNA-dependent RNA polymerase (RdRp). The function of other domains such as the Y domain, papain-like cysteine protease (PCP), and hypervariable region (HVR) is still undetermined [16][17].
ORF2 encodes for the capsid protein and has its role in HEV infection diagnostics and vaccine development. It is a 72 kDa protein that is N-glycosylated in mammalian cells [17][18].
ORF3 codes a small phosphoprotein (VP13) of 13 kDa and is phosphorylated at serine residue [16][18]. The ORF2 and ORF3 proteins are the results of subgenomic RNA translation [19].
ORF4 is unique to the HEV-1 genotype. It is entirely within ORF1 and overlaps with the X domain and helicase. Transcription of ORF4 protein is controlled by an internal ribosome entry site (IRES)-like RNA structure, and it plays a vital role in HEV RNA polymerase’s proper functioning. The ORF4 protein promotes viral RdRp activity and viral replication [14][18][20].
Hepatocytes are target cells where HEV replication takes place, as shown in Figure 2. HEV binds to the cell membrane through interactions with heparan sulfate proteoglycans, and enters the cell via clathrin-dependent endocytosis [21][22][23]. The virus genome is released into the cytoplasm, where viral RNA replication ensues. Positive-strand genomic RNA of 7.2 kb acts as messenger RNA for the translation of nonstructural ORF1 proteins. Based on positive-strand genomic viral RNA, a negative-strand RNA is synthesized, which subsequently serves as a template for synthesizing positive-strand 7.2 kb viral RNA and 2.2 kb subgenomic RNA [20][21][24]. Subgenomic RNA acts as a mRNA for the translation of ORF2 and ORF3 proteins. Translation of the capsid proteins and phosphoproteins occurs on the ribosomes of the host cell [1][19][23][24]. The next step is to assemble the virus and release the newly produced virions. Packaging of the viral genome in the capsid occurs spontaneously by the assembly of the non-glycosylated capsid proteins. The formation of a quasi-enveloped particle involves the phosphorylation of the VP13 protein. Virion particles become intracellular vesicles by the interaction of VP13 proteins with cell sorting host proteins. They are then released from the cell and can readily enter the bloodstream [11][19][20][21][25].
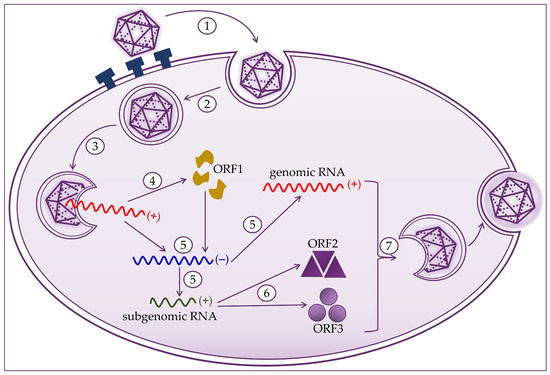
Figure 2. Hepatitis E virus life cycle consists of (1) attachment to heparan sulfate proteoglycans and entry to host cell; (2) clathrin-dependent endocytosis; (3) release of the positive-strand RNA genome into the cytosol; (4) translation to produce the ORF1 proteins; (5) replication via a negative-strand RNA mediator and synthesis of genomic and 2.2 kb subgenomic RNAs; (6) translation of the subgenomic RNA to produce the ORF2 and ORF3 proteins; and (7) packaging, virion assembly, and release of the newly formed virus.
3. Epidemiology of HEV
HEV is a major global health problem in developing and developed countries and is mostly zoonotic [11][26][27]. The World Health Organization (WHO) estimates that the incidence of HEV infections is 20 million annually, leading to approximately 3.3 million symptomatic cases of hepatitis E [28]. In 2015, it caused about 44,000 deaths, representing 3.3% of deaths due to viral hepatitis [28][29].
In the last two decades, the view that hepatitis E is a self-limiting tropical disease without a significant presence in developed countries has been revised because HEV has been found to circulate in Western countries [30][31]. It was noticed that HEV’s epidemiology is more complicated than it was initially thought. Namely, there are two different epidemiological forms of HEV [32][33][34]. Outbreaks of HEV-1 and HEV-2 occur periodically in several regions of Asia, Africa, Mexico, and the Middle East due to floods causing water supplies contamination with feces. The first HEV epidemic (1955–1956) infected 29,300 people in India [14][35][36]. In hyperendemic regions of the world, the infection is caused by HEV genotypes 1 and 2, most often in young adults, and extremely severe forms develop in pregnant women [17][37][38].
Genotype 1 causes infections in Asia (south, southeast, and central) and North Africa [17][39]. Furthermore, the presence of the HEV-1 genotype was observed in Cuba and Venezuela [32]. The HEV-1 genotype is a common cause of acute hepatitis E in Europe, introduced by international travelers, mostly from Asia [32]. The HEV-2 genotype was first detected in an epidemic in Mexico but was later seen in African countries (Chad, Central African Republic, Egypt, Democratic Republic of Congo, Nigeria, and Namibia) [40]. HEV-3 and HEV-4 infections are endemic in developed countries. They are mainly transmitted from animals or occur due to consuming contaminated food, and parenteral transmission via blood transfusion has been reported [11][13][41]. Genotype 3 is common among pigs worldwide, and they are a significant reservoir for human infections [13][42][43]. It has also been found in shellfish in Scotland and southern Italy [44]. Strains within genotype 4 are similar to genotype 3 [32], and they are widespread in South and East Asia [13].
The prime reservoir of hepatitis E are domestic pigs and wild boars [44]. Infections are more common in people over the age of 50, and recent studies have shown that infections are more common in men than in women [13][45]. They cause sporadic acute cases of hepatitis E in the United States, Europe, China, and Japan [40]. Genotypes 5 and 6 were recorded only in wild boars, and genotypes 7 and 8 were identified in camels and camelids [6], as shown in Table 2.
Table 2. Distribution of HEV genotypes.
|
HEV Genotype |
Host |
Endemic Regions |
|---|---|---|
|
1 |
Human |
Asia, North Africa, Mexico, Middle East |
|
2 |
Human |
Asia, North Africa, Mexico, Middle East |
|
3 |
Human, rabbit, pig, mongoose |
Worldwide |
|
4 |
Human, domestic pig, and wild boar |
South and East Asia, the United States, Europe, China, and Japan |
|
5 |
Wild boar |
Japan |
|
6 |
Wild boar |
Japan |
|
7 |
Camel |
Dubai |
|
8 |
Camel |
Dubai, western China |
It is estimated that HEV infections caused 70,000 deaths due to acute liver failure and about 3000 stillbirths [17]. China is considered an endemic HEV region [46]. Contact with cats and pigs, age, and soil exposure is associated with HEV infection; furthermore, studies have shown a high prevalence of HEV and significant potential for HEV infection transmission in pregnant women in China [47]. In the last ten years, more than 21,000 acute clinical cases of hepatitis E have been reported in European countries, with 28 deaths. The majority (80%) of patients are in Germany, France, and the United Kingdom [48]. The prevalence of HEV antibodies in England was 13%, increased over the years, and reached 25% in individuals over 50 years of age. Based on the incidence assessment, it has been proven that common risk factors still exist for acquiring HEV infection in all age groups in England. Current estimates of HEV incidence revealed that the incidence did not differ in different age groups [49]. Based on research conducted in England, excessive alcohol consumption and diabetes have been observed among people with acute hepatitis E, which are risk factors for liver fibrosis and steatosis [50].
Hepatitis E is a proven autochthonous zoonosis in Croatia caused by Orthohepevirus A genotype 3 (HEV-3) [51]. In Croatia, a seroprevalence of 32.94% in domestic pigs was found in 11 of 14 counties. With seropositive wild boars found in six of the 16 counties, the seroprevalence was 31.10%. The highest seroprevalence was found in Vukovar-Srijem and Osijek-Baranja counties (eastern Croatia), where pig breeding dominates and where wild boars’ highest density was recorded [52][53]. It has been proven that all detected HEV strains in Croatia are genetically closely related to strains found in humans and/or animals from other European countries. All of the above indicates that live animals’ trafficking or the wild boars’ movement increases HEV infection risk [54].
References
- LeDesma, R.; Nimgaonkar, I.; Ploss, A. Hepatitis E Virus Replication. Viruses 2019, 11, 719.
- Emerson, S.U.; Purcell, R.H. Hepatitis E virus. Rev. Med. Virol. 2003, 13, 145–154.
- Lhomme, S.; Marion, O.; Abravanel, F.; Izopet, J.; Kamar, N. Clinical Manifestations, Pathogenesis and Treatment of Hepatitis E Virus Infections. J. Clin. Med. 2020, 9, 331.
- Sridhar, S.; Teng, J.L.L.; Chiu, T.H.; Lau, S.K.P.; Woo, P.C.Y. Hepatitis E virus genotypes and evolution: Emergence of camel hepatitis E variants. Int. J. Mol. Sci. 2017, 18, 869.
- Chaudhry, S.A.; Verma, N.; Koren, G. Hepatitis E infection during pregnancy. Can. Fam. Physician 2015, 61, 607–608.
- Fousekis, F.S.; Mitselos, I.V.; Christodoulou, D.K. Extrahepatic manifestations of hepatitis E virus: An overview. Clin. Mol. Hepatol. 2020, 26, 16–23.
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Kumar, C.T.R.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521.
- Meng, X.J. Recent advances in Hepatitis E Virus. J. Viral Hepat. 2010, 17, 153–161.
- Goel, A.; Aggarwal, R. Hepatitis E: Epidemiology, Clinical Course, Prevention, and Treatment. Gastroenterol. Clin. N. Am. 2020, 49, 315–330.
- Yin, X.; Feng, Z. Hepatitis E Virus Entry. Viruses 2019, 11, 883.
- Wu, C.; Wu, X.; Xia, J. Hepatitis E virus infection during pregnancy. Virol. J. 2020, 17, 1–11.
- Al-Sadeq, D.W.; Majdalawieh, A.F.; Mesleh, A.G.; Abdalla, O.M.; Nasrallah, G.K. Laboratory challenges in the diagnosis of hepatitis E virus. J. Med. Microbiol. 2018, 67, 466–480.
- Pallerla, S.R.; Harms, D.; Johne, R.; Todt, D.; Steinmann, E.; Schemmerer, M.; Wenzel, J.J.; Hofmann, J.; Shih, J.W.K.; Wedemeyer, H.; et al. Hepatitis E Virus Infection: Circulation, Molecular Epidemiology, and Impact on Global Health. Pathogens 2020, 9, 856.
- Aslan, A.T.; Balaban, H.Y. Hepatitis E virus: Epidemiology, diagnosis, clinical manifestations, and treatment. World J. Gastroenterol. 2020, 26, 5543–5560.
- Lhomme, S.; Legrand-Abravanel, F.; Kamar, N.; Izopet, J. Screening, diagnosis and risks associated with Hepatitis E virus infection. Expert Rev. Anti-Infect. Ther. 2019, 17, 403–418.
- Khudyakov, Y.; Kamili, S. Serological diagnostics of hepatitis E virus infection. Virus Res. 2011, 161, 84–92.
- Kar, P.; Karna, R. A Review of the Diagnosis and Management of Hepatitis E. Curr. Treat. Options Infect. Dis. 2020, 12, 310–320.
- Nan, Y.; Zhang, Y.J. Molecular biology and infection of hepatitis E virus. Front. Microbiol. 2016, 7, 1419.
- Oechslin, N.; Moradpour, D.; Gouttenoire, J. On the Host Side of the Hepatitis E Virus Life Cycle. Cells 2020, 9, 1294.
- Kenney, S.P.; Meng, X.J. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724.
- Himmelsbach, K.; Bender, D.; Hildt, E. Life cycle and morphogenesis of the hepatitis E virus. Emerg. Microbes Infect. 2018, 7, 196.
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on hepatitis E virology: Implications for clinical practice. J. Hepatol. 2016, 65, 200–212.
- Ahmad, T.; Nasir, S.; Musa, T.H.; AlRyalat, S.A.S.; Khan, M.; Hui, J. Epidemiology, diagnosis, vaccines, and bibliometric analysis of the 100 top-cited studies on Hepatitis E virus. Hum. Vaccines Immunother. 2020, 1–15.
- Panda, S.K.; Varma, S.P.K. Hepatitis E: Molecular Virology and Pathogenesis. J. Clin. Exp. Hepatol. 2013, 3, 114–124.
- Ju, X.; Ding, Q. Hepatitis E virus assembly and release. Viruses 2019, 11, 539.
- Thakur, V.; Ratho, R.K.; Kumar, S.; Saxena, S.K.; Bora, I.; Thakur, P. Viral Hepatitis E and Chronicity: A Growing Public Health Concern. Front. Microbiol. 2020, 11.
- Sayed, I.M.; Vercouter, A.S.; Abdelwahab, S.F.; Vercauteren, K.; Meuleman, P. Is hepatitis E virus an emerging problem in industrialized countries? Hepatology 2015, 62, 1883–1892.
- World Health Organization Hepatitis, E. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-e (accessed on 17 December 2020).
- Teshale, E. Travel-Related Infectious Diseases. In Yellow Book; Brunette, G., Nemhauser, J., Eds.; Oxford University Press: New York, NY, USA, 2019.
- Horvatits, T.; Schulze zur Wiesch, J.; Lütgehetmann, M.; Lohse, A.W.; Pischke, S. The Clinical Perspective on Hepatitis E. Viruses 2019, 11, 617.
- Denner, J. Hepatitis e virus (HEV)—The future. Viruses 2019, 11, 251.
- Clemente-Casares, P.; Ramos-Romero, C.; Ramirez-Gonzalez, E.; Mas, A. Hepatitis E virus in industrialized countries: The silent threat. Biomed. Res. Int. 2016, 2016.
- Primadharsini, P.P.; Nagashima, S.; Okamoto, H. Genetic variability and evolution of hepatitis E virus. Viruses 2019, 11, 456.
- Gupta, E.; Agarwala, P. Hepatitis E virus infection: An old virus with a new story! Indian J. Med. Microbiol. 2018, 36, 317–323.
- Hakim, M.S.; Wang, W.; Bramer, W.M.; Geng, J.; Huang, F.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. The global burden of hepatitis E outbreaks: A systematic review. Liver Int. 2017, 37, 19–31.
- Teshale, E.H.; Hu, D.J.; Holmberg, S.D. The two faces of hepatitis E virus. Clin. Infect. Dis. 2010, 51, 328–334.
- Narayanan, S.; Abutaleb, A.; Sherman, K.E.; Kottilil, S. Clinical features and determinants of chronicity in hepatitis E virus infection. J. Viral Hepat. 2019, 26, 414–421.
- Sooryanarain, H.; Meng, X.J. Hepatitis E virus: Reasons for emergence in humans. Curr. Opin. Virol. 2019, 34, 10–17.
- Blasco-Perrin, H.; Abravanel, F.; Blasco-Baque, V.; Péron, J.M. Hepatitis E, the neglected one. Liver Int. 2016, 36, 130–134.
- Carratalà, A.; Joost, S. Population density and water balance influence the global occurrence of hepatitis E epidemics. Sci. Rep. 2019, 9.
- Wallace, S.J.; Crossan, C.; Hussaini, S.H.; Dalton, H.R. Hepatitis E: A largely underestimated, emerging threat. Br. J. Hosp. Med. 2019, 80, 399–404.
- Wedemeyer, H.; Pischke, S.; Manns, M.P. Pathogenesis and treatment of hepatitis E virus infection. Gastroenterology 2012, 142.
- Sclair, S.N.; Schiff, E.R. An update on the hepatitis E virus. Curr. Gastroenterol. Rep. 2013, 15.
- Takova, K.; Koynarski, T.; Minkov, I.; Ivanova, Z.; Toneva, V.; Zahmanova, G. Increasing Hepatitis E Virus Seroprevalence in Domestic Pigs and Wild Boar in Bulgaria. Animals 2020, 10, 1521.
- Khuroo, M.S.; Khuroo, M.S.; Khuroo, N.S. Hepatitis E: Discovery, global impact, control and cure. World J. Gastroenterol. 2016, 22, 7030–7045.
- Wang, Y.; Wang, S.; Wu, J.; Jiang, Y.; Zhang, H.; Li, S.; Liu, H.; Yang, C.; Tang, H.; Guo, N.; et al. Hepatitis E virus infection in acute non-traumatic neuropathy: A large prospective case-control study in China. EBioMedicine 2018, 36, 122–130.
- Cong, W.; Sui, J.C.; Zhang, X.Y.; Qian, A.D.; Chen, J.; Zhu, X.Q. Seroprevalence of hepatitis E virus among pregnant women and control subjects in China. J. Med. Virol. 2015, 87, 446–450.
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernandez Escamez, P.S.; Herman, L.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; et al. Public health risks associated with hepatitis E virus (HEV) as a food-borne pathogen. EFSA J. 2017, 15.
- Ijaz, S.; Vyse, A.J.; Morgan, D.; Pebody, R.G.; Tedder, R.S.; Brown, D. Indigenous hepatitis E virus infection in England: More common than it seems. J. Clin. Virol. 2009, 44, 272–276.
- Webb, G.W.; Dalton, H.R. Hepatitis E: An underestimated emerging threat. Ther. Adv. Infect. Dis. 2019, 6.
- Đaković Rode, O.; Jemeršić, L.; Vince, A. Hepatitis E in Croatia-Guidelines for Diagnosis and Treatment. Liječnički Vjesn. 2016, 138, 289–296.
- Jemeršić, L.; Keros, T.; Maltar, L.; Barbić, L.; Čavlek, T.V.; Jeličić, P.; Rode, O.Đ.; Prpić, J. Differences in hepatitis E virus (HEV) presence in naturally infected seropositive domestic pigs and wild boars—An indication of wild boars having an important role in HEV epidemiology. Vet. Arh. 2017, 87, 651–663.
- Mrzljak, A.; Dinjar-Kujundzic, P.; Jemersic, L.; Prpic, J.; Barbic, L.; Savic, V.; Stevanovic, V.; Vilibic-Cavlek, T. Epidemiology of hepatitis e in South-East Europe in the “one Health” concept. World J. Gastroenterol. 2019, 25, 3168–3182.
- Jemeršić, L.; Prpić, J.; Brnić, D.; Keros, T.; Pandak, N.; Rode, O.Đ. Genetic diversity of hepatitis E virus (HEV) strains derived from humans, swine and wild boars in Croatia from 2010 to 2017. BMC Infect. Dis. 2019, 19, 269.





