
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marta Palusinska-Szysz | + 2834 word(s) | 2834 | 2021-02-17 06:29:13 | | | |
| 2 | Vicky Zhou | Meta information modification | 2834 | 2021-02-28 04:17:19 | | |
Video Upload Options
Legionella are Gram-stain-negative rods associated with water environments: either natural or man-made systems. The inhalation of aerosols containing Legionella bacteria leads to the development of a severe pneumonia termed Legionnaires’ disease.
1. Introduction
Bacteria from the family Legionellaceae are Gram-negative bacilli, which are part of the natural microflora of aquatic and soil environments, as well as anthropogenic ecosystems. From the microbiological point of view, the ecology of Legionella bacilli is highly complex. Legionella bacteria survive in a temperature range between 5 °C and 65 °C and at pH 7–9; however, due to their specific nutritional requirements and a narrow range of environmental conditions necessary for their growth, they are unable to compete with other bacteria [1]. The bacteria require specific physicochemical conditions for development, which can only be fulfilled inside the eukaryotic host. Protozoa, commonly found in natural ecosystems, are an important link in the food chain and exert a significant impact on bacterial populations. However, not all bacteria are ingested, killed, and digested by protozoa. The outcome of the Legionella–protozoa interaction is associated with the host species. In some cases, Legionella resists digestion and kills the host, and in others, the protist digests the bacterium [2]. A total of 61 different Legionella species and more than 70 serogroups have been identified [3]. An in-silico analysis of the Legionella pneumophila genome identified several genomic islands indispensable for bacterial growth within protozoa, which differ among the various amoeba species [4]. Free-living amoebae from the genus Acanthamoeba, Naegleria, and Vermamoeba (Hartmanella) and ciliates from the genus Tetrahymena provide an intracellular niche in which Legionella can proliferate [5][6]. The intracellular growth of the bacteria has been associated with the enhanced environmental survival, virulence, and antibiotic resistance of the bacteria [7][8]. L. pneumophila, i.e., the most pathogenic species of this family, use many molecular and cellular aspects of intracellular proliferation inside protozoa in patho-adaptation to human cells [9]. Legionella enter the human organism from the natural environment via contaminated aerosols generated by cooling towers, large air-conditioning systems, fountains, showers, groundwater used for sprinkler irrigation, and similar sources [10][11]. This mode of bacterial spread is regarded as the major infection route, although bacterial transmission from person to person has been reported as well [12]. In the human organism, the bacteria cause respiratory infections with varying severity: from a flu-like infection called Pontiac fever, which does not require specialised treatment, to an acute, multi-lobar pneumonia called Legionnaires’ disease, which may result in death [13]. The bacteria proliferate robustly in lung alveolar macrophages, leading to tissue damage and the subsequent development of the disease. The macrophage resistance of Legionella spp. is a prerequisite for their virulence.
Bacterial survival in contact with eukaryotic cells—in particular, those predisposed for killing bacteria—depends on many determinants of adaptation to an environment that is extremely hostile but rich in nutrients. The bacteria overcome the killing mechanisms of phagocytes by means of specialised protein translocation systems: type II Lsp and type IVB Dot/Icm. The type II system controls the secretion of enzymes (lipases, protease, and RNase), thus promoting growth, intracellular replication, and virulence [14]. The Dot/Icm transporter delivers substrates that modulate multiple host cell processes, resulting in the biogenesis of the L. pneumophila-containing vacuole (LCV) permissive for intracellular bacterial replication. L. pneumophila achieve host infection by exporting approximately 300 substrates of the Dot/Icm system across one or two cell membranes to the site of action [15]. The precise delivery of the effectors to the host cell in a timely manner and space is possible thanks to an efficiently functioning bacterial cell envelope. The cell envelope of L. pneumophila is typical for Gram-negative bacteria and consists of two distinct membranes, the inner (IM) and outer membrane (OM), separated by the periplasm. The periplasm contains a relatively thin layer of strongly crosslinked peptidoglycan and various proteins. Peptidoglycan is composed of muramic acid, glucosamine, glutamic acid, alanine, and meso-diaminopimelic acid in a molar ratio of 0.8:0.8:1.1:1.7:1 [16]. The OM is asymmetric, with an inner leaflet mostly composed of phospholipids and an outer leaflet mostly comprising lipopolysaccharides (LPS). This asymmetry is critical for maintaining the OM permeability barrier. LPS consists of three regions: O-antigen, core, and lipid A. The lipid A region anchors LPS molecules to the outer membrane through hydrophobic interactions with the acyl chains of the phospholipids (PLs) constituting the inner layer of this membrane [17]. In addition to proteins, which have a fundamental importance for various aspects of cell physiology, including the infection of a host organism, lipids are involved in the highly specific interactions with the host cell. Three types of lipid-containing molecules are present in the cell envelope of L. pneumophila: phospholipids (PL), lipopolysaccharide (LPS), and lipoproteins (Figure 1).
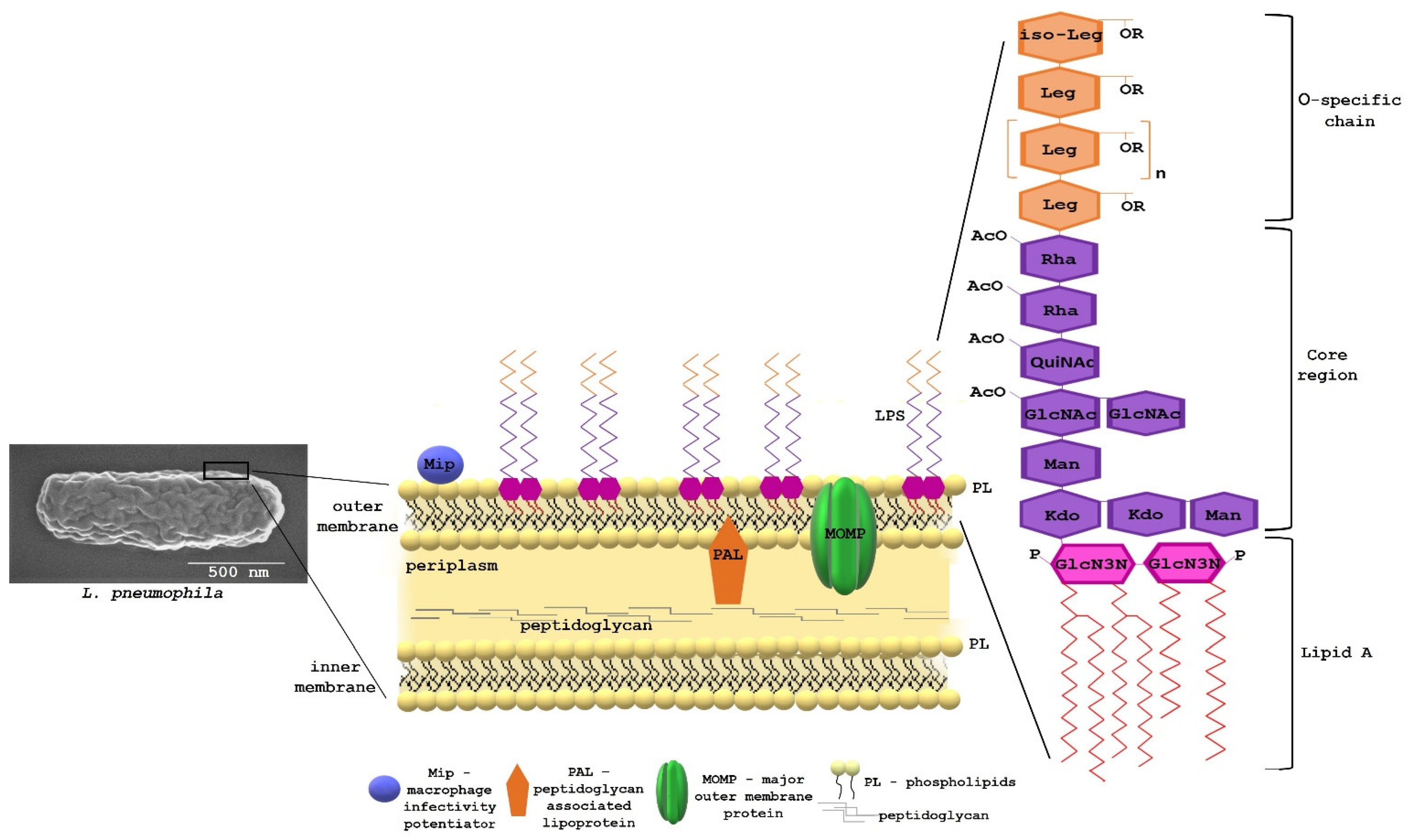
2. Biological Significance of Legionella LPS
L.pneumophila LPS is the main antigen recognised by antibodies contained in the serum of patients and the thermostable antigen excreted in urine [18]. The urinary antigen test is the most commonly used method for the diagnosis of Legionnaires’ disease [19]. The use of monoclonal antibodies (mAb) of the Dresden panel, recognising epitopes located in the highly heterogeneous O-specific chain of L. pneumophila LPS, facilitated distinguishing 15 serogroups and nine subgroups within serogroup 1 [20]. The two antibodies (mAb3/1 and mAb8/5) that recognise the virulent strains responsible for the majority of laboratory-confirmed cases of Legionnaires’ disease turned out to be valuable in epidemiological studies and for clinical purposes. The characterisation of the LPS biosynthesis loci of L. pneumophila serogroup 1 strains revealed two major regions: a specific 18-kb region and a conserved 15-kb region containing genes found in serogroup 1 and non-serogroup 1 strains. The most variable region is involved in O-antigen modification [21]. Antibody mAb3/1 recognises an epitope associated with the 8-O-acetyl group in legionaminic acid. The O-acetyl-transferase enzyme encoded by the lag-1 gene is responsible for the transfer of the O-acetyl group to legionaminic acid [22]. Studies carried out in Europe and Asia have shown that the lag-1 gene was harboured by a significantly higher number of clinical isolates of L. pneumophila sg1 compared with environmental isolates [23][24].
The LPS of L. pneumophila is highly hydrophobic due to the presence of deoxy groups and N- and O-acyl substituents in legionaminic acid, but the highest degree of hydrophobicity is exhibited by lag-1 strains producing 8-O-acetyl groups, which most likely contributes to the transmissibility of these bacteria in aerosols and adhesion to host cells. A TF3/1 mutant in the lag-1 gene defective in the synthesis of 8-O-acetyl substituents, not recognised by the mAb3/1 antibody, adhered less strongly to the macrophages of the THP-1 line and to A. castellanii cells, compared to the wild-type strain [25]. This mutant also failed to produce high-molecular-weight long-chain O-polysaccharide [26]. Comparative studies of the kinetics of interactions between the host cell and the L. pneumophila wild-type strain or the mutant showed a higher efficiency of binding to the amoeba surface in bacteria with the full-length O-chain and 8-O-acetyl groups. However, both strains multiplied inside the host cells successfully, irrespective of the differences in the length and structure of the polysaccharide part of LPS. Previous studies also showed that L. pneumophila strains lacking 8-O-acetyl substituents were as effective in infecting amoebae and macrophages as strains that expressed this LPS motif [26]. Thus, the LPS of L. pneumophila plays a critical role in the early stages of infection, anchoring the bacteria to the host cell’s membrane. Disturbances in the synthesis of the polysaccharide region of L. pneumophila LPS exert an effect on the composition and structure of phospholipids and proteins. The TF3/1 mutant showed differences in the structures of the PC and PG species, compared to the wild-type strain. Moreover, the mutant strain was synthesised by 11 mol% lower amounts of branched fatty acids and approximately two-fold higher amounts of long-chain fatty acid (20:0) than the wild-type strain. The changes in the surface components determined the cell surface topography of these bacteria and their nanomechanical properties. The TF3/1 mutant had grooves on the cell surface and did not produce as many outer membrane vesicles (OMV) as the wild-type strain [25]. Spherical bilayers consisting of LPS, phospholipids, outer membrane proteins, and periplasmic components are naturally secreted from the cell envelope of L. pneumophila. OMVs play a significant role in the pathogenesis of these bacteria, simultaneously delivering multiple virulence factors to host cells and tissues [27]. The release of the vesicles from L. pneumophila is developmentally regulated, i.e., the vesicles are connected to the cell wall but shed LPS in the replicative phase and are profusely released in the transmissive phase [28]. One of the functions of L. pneumophila OMVs is the inhibition of phagosome–lysosome fusion during the intracellular infection of macrophages. This capability was correlated with growth phase-dependent modifications of the composition of glycoconjugates contained in the vesicles. The LPS of the bacteria in the transmissive phase of growth is deacetylated and elongated to a form that effectively blocks the fusion between phagosomes and lysosomes and, thus, independently of the effectors of the IV secretion system, inhibits the maturation of macrophages [29]. In turn, during the replicative phase, the degree of acetylation of the LPS O-chain increases, which probably contributes to the increased tolerance to the hostile conditions of the intracellular environment of macrophages [29]. Seeger et al. showed that LPS fractions below 300 kDa, not associated with OMV, significantly delayed phagolysosomal maturation one hour after phagocytosis, regardless of the bacterial growth phase [30].
LPS expressed by L. pneumophila in the transmissive phase binds to a sialic acid-specific lectin more strongly than LPS from bacteria in the replicative phase of growth. The ability of the bacteria to bind to this receptor correlates with the effectiveness of macrophage infection [29]. Legionaminic acid of L. pneumophila sg1 shares the same D-glycero-D-galacto absolute configuration as 5-acetamido neuraminic acid (Neu5Ac, sialic acid). Neu5Ac, located on the surface of mammalian cells, is involved in cell–cell interactions and the immune response [31]. The molecular mimicry of eukaryotic cell macromolecules is one of the strategies used by Legionella bacteria to colonise host cells. The structural similarity of legionaminic and neuraminic acid may be an example of mimicry to the host cell used not only in the adhesion process but, also, in modulation of the immune response to infection. The LPS of L. pneumophila is less toxic and less potent in its ability to induce proinflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) from Mono Mac 6 cells, compared to the highly pyrogenic LPS from Enterobacteriaceae members [32]. The bioactive centre of LPS is lipid A, whose toxicity is directly influenced by the length and number of groups of the fatty acids attached to its glycosidic backbone. The presence of fatty acid chains twice the length of the corresponding chains found in the majority of toxic lipids A containing C12, C12OH, C14, and C14OH account for the low endotoxic activity of L. pneumophila LPS due to a failure to interact with receptor CD14 (a glycosylphosphatidylinositol-anchored protein) and with its soluble form [32]. Host pattern recognition receptors such as Toll-like receptors (TLRs) are involved in the process of recognition of LPS. TLR4 functions as a sensor of LPS on the OM in Gram-negative bacteria, promptly inducing the production of antibacterial cytokines. The lipid A region of L. pneumophila LPS was a weak TLR4 agonist [33]. Additionally, macrophages of TLR4-deficient mice infected by L. pneumophila were not defective in the production of cytokines, and the rate of bacterial clearance from the lungs of these mice was similar to that in wild-type mice [34][35]. However, the TLR2-deficient mice were more sensitive to L. pneumophila than TLR2-sufficient mice [35][36]. These results indicated that LPS from L. pneumophila was recognised by TLR2, which is a typical receptor for peptidoglycan [33]. This unusual detection pattern of switching TLR4 to TLR2 is related to the presence of substituent (a ketone group at C27) or a branch on the penultimate carbon of different fatty acids of L. pneumophila lipid A [33]. Upon recognition by TLR2, LPS triggers signalling pathways controlled by the MyD88 adaptor protein, which results in the production of inflammatory cytokines and subsequent clearance of L. pneumophila from the lungs [37]. The level of inflammatory cytokine production was reduced in TLR2 and MyD88 gene knockdown macrophages of the U937 cell line infected by L. pneumophila [38]. Additionally, human macrophages induced by L. pneumophila OMVs synthesised IL-8 relying on TLR2-dependent signalling pathways [27].
The structure of lipid A composed of GlcN3N and long-chain fatty acids also provides a protective mechanism against the degradation of lipid A/LPS by amidases and/or esterases of amoebae during the intracellular growth of L. pneumophila [39].
L. pneumophila LPS is subject to phase variations correlated with the attenuation of virulence traits such as the ability to multiply within macrophage-like HL60 cells or A. castellanii [40]. The molecular mechanism responsible for this variability consists of the chromosomal insertion and excision of an unstable 30-kb genetic element, which does not harbour genes related to LPS biosynthesis [41]. Phase variation in the RC1 strain of the L. pneumophila sg 1 subgroup OLDA influences the O-specific chain structure and the fatty acid profile of lipid A. The phase-variant strain 811 with the 29-kb element excised from the chromosome is devoid of N-methylation in legionaminic acid and contains shorter 3-hydroxy (16:0 and 18:0) fatty acids in lipid A [42][43].
L.pneumophila strain PtVFX/2014, associated with the first evidence of person-to-person transmission, i.e., a strain that was able to overcome the transmission barrier of human innate immunity, carried eight horizontally transferred regions encompassing genes involved in, e.g., LPS biosynthesis [44]. Epidemiological studies showed that the genes of the LPS cluster determining sg 1 of L. pneumophila were present in highly diverse genomic backbones of the strains responsible for the largest outbreaks of Legionnaires’ disease described so far and that it probably constitutes a major determinant of the human disease itself [44].
On the one hand, the LPS composed of three distinct regions in terms of structure and biological properties is an important factor in the virulence of Legionella bacteria, participating in complex mechanisms of the induction of lesions. On the other hand, LPS is a ligand recognised by proteins involved in the response to infection. During the infection of macrophages with L. pneumophila, host guanylate binding proteins (GBPs), triggered by cytoplasmic LPS derived from the bacteria, induce caspase-11-dependent pyroptosis [45][46]. The LPS of L. pneumophila is a ligand for collectins, which play an important role in the innate immunity of the lung. After the direct binding of LPS, hydrophilic proteins A and D promote the localisation of L. pneumophila in the acidic environment of the lysosomes, thus attenuating the intracellular multiplication of the phagocytosed bacteria [47]. Additionally, human apolipoprotein E binds to L. pneumophila LPS, which results in disturbances in the normal structure of the bacterial surface, and these changes may lead to impaired penetration of the host cells [48]. Two molecules of apoLp-III bind to a single micelle of L. dumoffii LPS formed from 12 to 29 monomeric LPS molecules, pointing to new strategies for anti-Legionella therapies [49].
Legionella spp. are important aetiological agents of pneumonia. They are responsible for 2–8% of community-acquired pneumonia cases [50]. A review of 46 CAP (community-acquired pneumonia) studies from European countries has indicated that Legionella spp. are particularly frequent among patients who require admission to an intensive care unit (ICU) [51]. In the USA, the reported cases of Legionnaires’ disease increased from 2.301 in 2005 to 7.104 in 2018. In Europe, the number of known cases of Legionnaires’ disease has almost doubled (from 1.3/100,000 people in 2014 to 2.3/100,000 people in 2018) [52]. The increase in cases may be related to the changing environmental conditions, which favour the growth of Legionella bacteria, as well as the increasing number of people susceptible to infection, such as the elderly and immunocompromised subjects. The closure of public buildings related to the SARS-CoV-2 pandemic has led to the long-term stagnation of water in installations and created optimal conditions for intensive multiplication of the bacteria, which substantially increased the risk of Legionella infections [53]. L. pneumophila produces a variety of cytosolic and cell envelope-associated lipids with a unique structure that plays an important role in its physiology and promotes bacterial adhesion and adaptation to the host’s intracellular environment.
Although more than 60 different Legionella species are known, with approximately half of the number of species isolated from clinical specimens, the complete structure of LPS is only known in L. pneumophila. The analysis of the genomes of various Legionella species has shown that these bacteria contain genes whose products are involved in the synthesis of lipids characteristic for eukaryotic cells, such as PC, or, e.g., hopanoids in L. falonii [54]. Lipid components, especially the unique ones, can serve as important biomarkers used in diagnostics and in taxonomic studies of this bacterial group. Bacterial lipid membranes are targets for the development of new antimicrobial drugs and a promising object of research in the fight against resistant bacteria [55]. Therefore, the elucidation of the complex lipid structure of the cell envelope of various Legionella spp. using advanced lipidomic technologies may be important for the development of new strategies for the prevention and treatment of Legionella infections.
References
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 2002, 15, 506–526.
- Amaro, F.; Wang, W.; Gilbert, J.A.; Anderson, O.R.; Shuman, H.A. Diverse protist grazers select for virulence-related traits in Legionella. ISME J. 2015, 9, 1607–1618.
- Euzeby, J.P. List of Prokaryotic Names with Standing in Nomenclature Genus Legionella. Available online: https://www.bacterio.net/legionella.html (accessed on 27 January 2021).
- O’Connor, T.J.; Adepoju, Y.; Boyd, D.; Isberg, R.R. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. USA 2011, 108, 14733–14740.
- Shaheen, M.; Ashbolt, N.J. Differential bacterial predation by free-living amoebae may result in blooms of Legionella in drinking water systems. Microorganisms 2021, 15, 174.
- Brieland, J.; McClain, M.; LeGendre, M.; Engleberg, C. Intrapulmonary Hartmannella vermiformis: A potential niche for Legionella pneumophila replication in a murine model of legionellosis. Infect. Immun. 1997, 65, 4892–4896.
- Personnic, N.; Striednig, B.; Lezan, E.; Manske, C.; Welin, A.; Schmidt, A.; Hilbi, H. Quorum sensing modulates the formation of virulent Legionella persisters within infected cells. Nat. Commun. 2019, 10, 5216.
- Brieland, J.; McClain, M.; Heath, L.; Chrisp, C.; Huffnagle, G.; LeGendre, M.; Hurley, M.; Fantone, J.C.; Engleberg, C. Coinoculation with Hartmanella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires’ disease. Infect. Immun. 1996, 64, 2449–2456.
- Al-Quadan, T.; Price, C.T.; Kwaik, Y.A. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 2012, 20, 299–306.
- De Giglio, O.; Fasano, F.; Diella, G.; Lopuzzo, M.; Napoli, C.; Apollonio, F.; Brigida, S.; Calia, C.; Campanale, C.; Marzella, A.; et al. Legionella and legionellosis in touristic-recreational facilities: Influence of climate factors and geostatistical analysis in Southern Italy (2001–2017). Environ. Res. 2019, 178, 108721.
- De Giglio, O.; Napoli, C.; Apollonio, F.; Brigida, S.; Marzella, A.; Diella, G.; Calia, C.; Scrascia, M.; Pacifico, C.; Pazzani, C.; et al. Occurrence of Legionella in groundwater used for sprinkler irrigation in Southern Italy. Environ. Res. 2019, 170, 215–221.
- Correia, A.M.; Ferreira, J.S.; Borges, V.; Nunes, A.; Gomes, B.; Capucho, R.; Gonçalves, J.; Antunes, D.M.; Almeida, S.; Mendes, A.; et al. Probable person-to-person transmission of Legionnaires’ disease. N. Engl. J. Med. 2016, 374, 497–498.
- Cunha, B.A.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385.
- Portlock, T.J.; Tyson, J.Y.; Dantu, S.C.; Rehman, S.; White, R.C.; McIntire, I.E.; Sewell, L.; Richardson, K.; Shaw, R.; Pandini, A.; et al. Structure, dynamics and cellular insight into novel substrates of the Legionella pneumophila type II secretion system. Front. Mol. Biosci. 2020, 11, 112.
- Bärlocher, K.; Welin, A.; Hilbi, H. Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Front. Cell. Infect. Microbiol. 2017, 7, 482.
- Shevchuk, O.; Jäger, J.; Steinert, M. Virulence properties of the Legionella pneumophila cell envelope. Front. Microbiol. 2011, 25, 74.
- Caroff, M.; Novikov, A. LPS structure, function, and heterogeneity. In Endotoxin Detection and Control in Pharma, Limulus, and Mammalian Systems; Williams, K.L., Ed.; Springer Nature: Berlin/Heidelberg, Germany, 2019; pp. 53–93.
- Ciesielski, C.A.; Blaser, M.J.; Wang, W.L. Serogroup specificity of Legionella pneumophila is related to lipopolysaccharide characteristics. Infect. Immun. 1986, 51, 397–404.
- Helbig, J.H.; Jacobs, E.; Lück, C. Legionella pneumophila urinary antigen subtyping using monoclonal antibodies as a tool for epidemiological investigations. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1673–1677.
- Joly, J.R.; McKinney, R.M.; Tobin, J.O.; Bibb, W.F.; Watkins, I.D.; Ramsay, D. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 1986, 23, 768–771.
- Petzold, M.; Thürmer, A.; Menzel, S.; Mouton, J.W.; Heuner, K.; Lück, C. A structural comparison of lipopolysaccharide biosynthesis loci of Legionella pneumophila serogroup 1 strains. BMC Microbiol. 2013, 13, 198.
- Zou, H.C.; Knirel, Y.A.; Helbig, H.J.; Zähringer, U.; Mintz, C.S. Molecular cloning and characterization of a locus responsible for O-acetylation of the O polysaccharide of Legionella pneumophila serogroup 1 lipopolysaccharide. J. Bacteriol. 1999, 181, 4137–4141.
- Helbig, J.H.; Lück, P.C.; Knirel, Y.A.; Witzleb, W.; Zähringer, U. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol. Infect. 1995, 115, 71–78.
- Jiang, L.; Amemura-Maekawa, J.; Ren, H.; Li, Y.; Sakata, M.; Zhou, H.; Murai, M.; Chang, B.; Ohnishi, M.; Qin, T. Distribution of lag-1 alleles, ORF7, and ORF8 genes of lipopolysaccharide and sequence-based types among Legionella pneumophila serogroup 1 isolates in Japan and China. Front. Cell. Infect. Microbiol. 2019, 9, 274.
- Palusinska-Szysz, M.; Luchowski, R.; Gruszecki, W.I.; Choma, A.; Szuster-Ciesielska, A.; Lück, C.; Petzold, M.; Sroka-Bartnicka, A.; Kowalczyk, B. The role of Legionella pneumophila serogroup 1 lipopolysaccharide in host-pathogen interaction. Front. Microbiol. 2019, 10, 2890.
- Lück, P.C.; Freier, T.; Steudel, C.; Knirel, Y.A.; Lûneberg, E.; Zähringer, U.; Helbig, J.H. A point mutation in the active site of Legionella pneumophila O-acetyltransferase results in modified lipopolysaccharide but does not influence virulence. Int. J. Med. Microbiol. 2001, 291, 345–352.
- Jäger, J.; Keese, S.; Roessle, M.; Steinert, M.; Schromm, A.B. Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell. Microbiol. 2015, 5, 607–620.
- Helbig, J.H.; Fernandez-Moreira, E.; Jacobs, E.; Luck, P.C.; Witt, M. Lipopolysaccharide architecture of Legionella pneumophila grown in broth and host cells. In Legionella: State of the Art 30 Years After its Recognition; Cianciotto, N.P., Ed.; ASM Press: Washington, DC, USA, 2006; pp. 261–264.
- Fernandez-Moreira, E.; Helbig, J.H.; Swanson, M.S. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect. Immun. 2006, 74, 3285–3295.
- Seeger, E.M.; Thuma, M.; Fernandez-Moreira, E.; Jacobs, E.; Schmitz, M.; Helbig, J.H. Lipopolysaccharide of Legionella pneumophila shed in a liquid culture as a nonvesicular fraction arrests phagosome maturation in amoeba and monocytic host cells. FEMS Microbiol. Lett. 2010, 307, 113–119.
- Ghosh, S. Sialic acid and biology of life: An introduction. In Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease, 1st ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–61.
- Neumeister, B.; Faigle, M.; Sommer, M.; Zähringer, U.; Stelter, F.; Menzel, R.; Schütt, C.; Northoff, H. Low endotoxic potential of Legionella pneumophila lipopolysaccharide due to failure of interaction with the monocyte lipopolysaccharide receptor CD14. Infect. Immun. 1998, 66, 4151–4157.
- Girard, R.; Pedron, T.; Uematsu, S.; Balloy, V.; Chignard, M.; Akira, S.; Chaby, R. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J. Cell Sci. 2003, 116, 293–302.
- Lettinga, K.D.; Florquin, S.; Speelman, P.; van Ketel, R.; van der Poll, T.; Verbon, A. Toll-like receptor 4 is not involved in host defense against pulmonary Legionella pneumophila infection in a mouse model. J. Infect. Dis. 2002, 186, 570–573.
- Archer, K.A.; Roy, C.R. MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires’ disease. Infect. Immun. 2006, 74, 3325–3333.
- Hawn, T.R.; Smith, K.D.; Aderem, A.; Skerrett, S.J. Myeloid differentiation primary response gene (88)-and toll-like receptor 2-deficient mice are susceptible to infection with aerosolized Legionella pneumophila. J. Infect. Dis. 2006, 193, 1693–1702.
- Archer, K.A.; Alexopoulou, L.; Flavell, R.A.; Roy, C.R. Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell. Microbiol. 2009, 11, 21–36.
- Mallama, C.A.; McCoy-Simandle, K.; Cianciotto, N.P. The type II secretion system of Legionella pneumophila dampens the MyD88 and Toll-like receptor 2 signaling pathway in infected human macrophages. Infect. Immun. 2017, 85, e00897-16.
- Zähringer, U.; Knirel, A.Y.; Lindner, B.; Helbig, J.H.; Sonesson, A.; Marre, R.; Rietschel, E.T. The lipopolysaccharide of Legionella pneumophila (strain Philadelphia 1): Chemical structure and biological significance. Prog. Clin. Biol. Res. 1995, 392, 113–139.
- Lüneberg, E.; Zähringer, U.; Knirel, Y.A.; Steinmann, D.; Hartmann, M.; Steinmetz, I.; Rohde, M.; Köhl, J.; Frosch, M. Phase-variable expression of lipopolysaccharide contributes to the virulence of Legionella pneumophila. J. Exp. Med. 1998, 188, 49–60.
- Lüneberg, E.; Mayer, B.; Daryab, N.; Kooistra, O.; Zähringer, U.; Rohde, M.; Swanson, J.; Frosch, M. Chromosomal insertion and excision of a 30 kb unstable genetic element is responsible for phase variation of lipopolysaccharide and other virulence determinants in Legionella pneumophila. Mol. Microbiol. 2001, 39, 1259–1271.
- Kooistra, O.; Lüneberg, E.; Knirel, Y.A.; Frosch, M.; Zähringer, U. N-Methylation in polylegionaminic acid is associated with the phase-variable epitope of Legionella pneumophila serogroup 1 lipopolysaccharide. Identification of 5-(N,N-dimethylacetimidoyl) amino and 5-acetimidoyl(N-methyl)amino-7-acetamido-3,5,7,9-tetradeoxynon-2-ulosonic acid in the O-chain polysaccharide. Eur. J. Biochem. 2002, 269, 560–572.
- Kooistra, O.; Knirel, Y.A.; Lüneberg, E.; Frosch, M.; Zähringer, U. Phase variation in Legionella pneumophila serogroup 1, subgroup OLDA, strain RC1 influences lipid A structure. In Proceedings of the 5th International Conference on Legionella, Ulm, Germany, 26–29 September 2000; Marre, R., Abu Kwaik, Y., Bartlett, C., Cianciotto, N.P., Fields, B.S., Frosch, M., Hacker, J., Lück, P.C., Eds.; ASM Press: Washington, DC, USA, 2002; pp. 68–73.
- Borges, V.; Nunes, A.; Sampaio, D.A.; Vieira, L.; Machado, J.; Simões, M.J.; Gonçalves, P.; Gomes, J.P. Legionella pneumophila strain associated with the first evidence of person-to-person transmission of Legionnaires’ disease: A unique mosaic genetic backbone. Sci. Rep. 2016, 6, 26261.
- Case, C.L.; Kohler, L.J.; Lima, J.B.; Strowig, T.; de Zoete, M.R.; Flavell, R.A.; Zamboni, D.S.; Roy, C.R. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl. Acad. Sci. USA 2013, 110, 1851–1856.
- Pilla, D.M.; Hagar, J.A.; Haldar, A.K.; Mason, A.K.; Degrandi, D.; Pfeffer, K.; Ernst, R.K.; Yamamoto, M.; Miao, E.A.; Coers, J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA 2014, 111, 6046–6051.
- Sawada, K.; Ariki, S.; Kojima, T.; Saito, A.; Yamazoe, M.; Nishitani, C.; Shimizu, T.; Takahashi, M.; Mitsuzawa, H.; Yokota, S.; et al. Pulmonary collectins protect macrophages against pore-forming activity of Legionella pneumophila and suppress its intracellular growth. J. Biol. Chem. 2010, 285, 8434–8443.
- Palusinska-Szysz, M.; Zdybicka-Barabas, A.; Cytryńska, M.; Wdowiak-Wróbel, S.; Chmiel, E.; Gruszecki, W.I. Analysis of cell surface alterations in Legionella pneumophila cells treated with human apolipoprotein E. Pathog. Dis. 2015, 73, 1–8.
- Palusińska-Szysz, M.; Zdybicka-Barabas, A.; Luchowski, R.; Reszczyńska, E.; Śmiałek, J.; Mak, P.; Gruszecki, W.I.; Cytryńska, C. Choline supplementation sensitizes Legionella dumoffii to Galleria mellonella apolipophorin III. Int. J. Mol. Sci. 2020, 21, 5818.
- Viasus, D.; Calatayud, L.; McBrown, M.V.; Ardanuy, C.; Carratalà, J. Urinary antigen testing in community-acquired pneumonia in adults: An update. Expert Rev. Anti-Infect. Ther. 2019, 17, 107–115.
- Welte, T.; Torres, A.; Nathwani, D. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax 2012, 67, 71–79.
- Nisar, M.A.; Ross, K.E.; Brown, M.H.; Bentham, R.; Whiley, H. Legionella pneumophila and protozoan hosts: Implications for the control of hospital and potable water systems. Pathogens 2020, 9, 286.
- Palazzolo, C.; Maffongelli, G.; D’Abramo, A.; Lepore, L.; Mariano, A.; Vulcano, A.; Bartoli, T.A.; Bevilacqua, N.; Giancola, M.L.; Di Rosa, E.; et al. Legionella pneumonia: Increased risk after COVID-19 lockdown? Italy, May to June 2020. Euro Surveill. 2020, 25, 1–3.
- Gomez-Valero, L.; Rusniok, C.; Carson, D.; Mondino, S.; Pérez-Cobas, A.E.; Rolando, M.; Pasricha, S.; Reuter, S.; Demirtas, J.; Crumbach, J.; et al. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc. Natl. Acad. Sci. USA 2019, 5, 2265–2273.
- Chatterjee, R.; Chowdhury, A.R.; Mukherjee, D.; Chakravortty, D. Lipid larceny: Channelizing host lipids for establishing successful pathogenesis by bacteria. Virulence 2021, 12, 195–216.




