| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nikolaos Labrou | + 2361 word(s) | 2361 | 2021-02-24 10:21:54 | | | |
| 2 | Nikolaos Labrou | Meta information modification | 2361 | 2021-02-26 15:34:48 | | | | |
| 3 | Bruce Ren | -21 word(s) | 2340 | 2021-03-01 06:52:13 | | | | |
| 4 | Bruce Ren | -21 word(s) | 2340 | 2021-03-01 07:00:40 | | |
Video Upload Options
Sirtuins (SIRTs) are nicotinamide adenine dinucleotide-dependent histone deacetylases that in-corporate complex functions in the mechanisms of cell physiology. Mammals have seven distinct members of the SIRT family (SIRT1-7), which play an important role in a well-maintained net-work of metabolic pathways that control and adapt the cell to the environment, energy availabil-ity and cellular stress. Until recently, very few studies investigated the role of SIRTs in modulating viral infection and progeny. Recent studies have demonstrated that SIRT1 and SIRT2 are promis-ing antiviral targets because of their specific connection to numerous metabolic and regulatory processes affected during infection.
1. Introduction
Human SIRTs form an evolutionarily conserved family of proteins that are ubiquitously expressed in all taxa. SIRTs are classified as NAD+ dependent deacylases/mono-ADP ribosyltransferases that regulate numerous cellular functions, including metabolism, cell cycle, stress and longevity [1][2][3][4]. Their name comes from the first member of the family that was studied in Saccharomyces cerevisiae, which was characterized as “silent information regulator 2” (Sir2), because it appeared to be involved in gene transcription silencing [5]. Subsequent studies have confirmed that the enzymatic activity of SIRTs affects the expression of various genes through the deacetylation of the ε-terminus of the amino acid Lys residues of histones. The discovery of SIRT isoforms in yeast and later in bacteria, plants and mammals suggests that Sir2 belongs to a very large and ancient family of genes [6][7].
Early studies have established that SIRTs affect the lifespan of organisms. Indeed, not only in yeast but also in other organisms, such as Drosophila melanogaster and Caenorhabditis elegans, expression of SIRTs leads to a longer shelf life, following a mechanism similar to that of calorie restriction [8][9][10]. Under calorie restriction conditions, SIRTs are able to increase mitochondrial biogenesis, enhance metabolism [11] and reduce the levels of reactive oxygen species (ROS), thus alleviating the progression of inflammation [12][13][14][15][16]. These observations opened new avenues in understanding the mechanisms that are affected by the action of SIRTs.
Mammals have seven distinct members of the SIRT family, SIRT1-7, which play an important role in the regulation of gene expression, mainly by controlling the posttranslational modification status of histones and transcription factors [17][18]. Another role that has been assigned to SIRTs is the regulation of a well-maintained network of metabolic pathways [17][18][19]. SIRTs were originally classified as NAD+-dependent histone deacetylases, indicating that these enzymes are involved in different metabolic pathways as part of the transcriptional adaptation mechanisms, acting as metabolic sensors. For example, the three mitochondrial SIRTs (SIRT3, SIRT4 and SIRT5) can control essential metabolic pathways. SIRT3 affects a range of key metabolic processes, such as fatty acid oxidation and the tricarboxylic acid (TCA) cycle [13]. SIRT4 inhibits the pyruvate dehydrogenase complex [20]. Furthermore, SIRT5 with its desuccinylase and demalonylase activity controls several metabolic pathways, including the urea cycle [21][22][23]. Considering their role in the regulation of metabolism, SIRTs were studied in the context of metabolic diseases [24][25][26][27][28]. Therefore, SIRTs regulate a large number of pathways, such as glucose and fatty acid metabolism, apoptosis, DNA repair, neuronal generation, inflammatory response and even the regulation of the circadian clock of organisms.
2. Acetylation and Deacetylation of Proteins
The conjugation of acetyl groups to proteins is a posttranslational modification mechanism that is widely used by eukaryotic cells to regulate various functions [29]. It is estimated that more than 6800 known acetylation sites exist in mammalian proteins [30]. A common and possibly more important form of acetylation is carried out post-translationally, at the ε-terminus amino group of Lys residues where an acetyl group is transferred from acetyl-CoA [31].
Lysine is a positively charged amino acid; therefore, its acetylation neutralize its positive charge and as a consequence alters the electrostatic properties of the protein [32][33]. This modification is a reversible process and takes place largely in histones, which are proteins that bind to DNA and form the basic structure of chromatin, the nucleosome. The group of enzymes that are responsible for the acetylation of histones are the histone acetyltransferase (HATs) [34]. The DNA has a negative charge while histones are rich in positively charged amino acid residues (lysine, arginine and histidine). The replication and transcription processes of DNA require the loss of interaction between DNA and histones, which is achieved by reducing the positive charge of histones. This is a reversible process, and therefore the degree of histone acetylation determines their access to DNA [32][33]. Conversely, histone deacetylation leads to transcriptional silencing of genes, as a heterochromatin environment is created around the gene, making it inaccessible to transcriptional mechanisms [35][36].
3. The Function, Structure and Regulation of SIRTs
3.1. The Human SIRT Family and Their Subcellular Status
SIRT genes are found in virtually all organisms, both prokaryotic (mycobacteria, eubacteria and archaea) and eukaryotic (yeasts and protozoa). In prokaryotes, SIRTs are encoded primarily by a single gene, while in eukaryotes by multiple genes. The variety of genes encoding these enzymes leads to the formation of isoforms, which have distinct catalytic functions and are localized in different subcellular compartments [37][38][39][40]. In mammals, the SIRT family consists of seven isoenzymes, SIRT1-7 (Table 1). Phylogenetic analysis of different SIRT genes (eukaryotic and prokaryotic) indicated that the seven mammalian SIRTs can be grouped into four different classes (I—IV) (Table 1). In Class I belongs the SIRT1, SIRT2 and SIRT3 and is divided in three sub-classes: a, b and c. Class II includes SIRT4, which also includes SIRTs from bacteria, insects, nematodes, fungus and protozoans. SIRT5 is part of the Class III SIRTs, whereas Class IV includes SIRT6 and SIRT7, divided in two sub-classes IVa and IVb
.
Table 1. The class, functional activity, molecular mass and chromosomal location of seven human SIRTs (SIRT1–7).
|
Sirtuins |
Class |
Functional Activity |
Molecular Mass (kDa) |
Chromosomal Location |
References |
|
SIRT1 |
I |
Deacetylase, Deacylase |
81,7 |
10q21.3 |
|
|
SIRT2 |
I |
Deacetylase, Deacylase |
41,5 |
19q13.3 |
|
|
SIRT3 |
I |
Deacetylase, Decrotonylase |
43.6 |
11p15.5 |
|
|
SIRT4 |
II |
Deacetylase, ADP-ribosyltransferase, Lipoamidase, Deacylase |
35.2 |
12q |
[49] |
|
SIRT5 |
III |
Deacetylase, Desuccinylase, Demalonylase, Deglutarylase |
33.9 |
6p23 |
|
|
SIRT6 |
IV |
Deacetylase, Demyristoylase, ADP-ribosyltransferase, Deacylase |
39.1 |
19p13.1 |
|
|
SIRT7 |
IV |
Deacetylase, Desuccinylase, |
44.9 |
17q25 |
[56] |
Each of SIRT isoenzymes is located in different subcellular compartments, depending on the function it performs, as shown in Figure 1. Some SIRTs can delocalize depending on the cell or tissue type, the developmental stage, metabolic status, and certain stress conditions. The distribution of SIRTs in different subcellular compartments allows them to interact with a wide variety of transcription factors and to participate in the regulation of different metabolic processes, such as apoptosis, glucose homeostasis, insulin resistance, stress resistance, circadian rhythm, mitochondrial biogenesis, and DNA repair. In addition, they play a key role in the development of inflammation and autophagy [56].
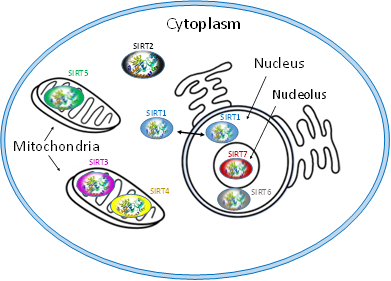
Figure 1. Subcellular location of seven mammalian SIRTS (SIRT1-7).
The isoenzymes SIRT1, SIRT6 and SIRT7 are found in the nucleus of the cell and epigenetically affect gene regulation, exerting their enzymatic activity as histone deacetylases. SIRT1 contains the nuclear localization signal peptide sequence (KRKKRK) in 41–46 residues; however, under certain conditions, it can also be transported from the nucleus to the cytoplasm where it is involved in the regulation of insulin metabolism. SIRT6 is found in heterochromatin and is involved in DNA repair mechanisms and also occurs in nucleoli during the G1 cell cycle. Overexpression of SIRT6 leads to slower mitotic process . SIRT6 is also localized in the endoplasmic reticulum, where it deacetylates tumor necrosis factor-α (TNF-α). SIRT7 is located at the nucleolus and is involved in rRNA transcription mechanism. The main site of SIRT2 is the cytoplasm, but in some phases of the cell cycle it is also found in the nucleus. SIRT2 is responsible for the deacetylation of tubulin microtubules and appears to play a key role in adipocyte differentiation. In addition, its action is necessary for the proper separation of chromosomes during mitosis as well as for the exit of cells from this phase [57].
The isoenzymes SIRT3, SIRT4 and SIRT5 are found in the mitochondria and contribute to oxidative stress alleviation by regulating the activity of specific metabolic enzymes and ATP synthesis, metabolism and intracellular signaling. SIRT3 can be moved between the nucleus and mitochondria under cellular stress.
3.2. Structure and Substrates of SIRTs
Numerous studies have examined the structural features of SIRTs (Figure 2) [57][58]. SIRTs composed by approximately 275 amino acids that are organized into two structurally distinct domains: the conserved large domain that has the characteristic structure of the Rossmann fold and the small domain that is less conserved among the members of the SIRT family and contains a zinc ion binding site with the consensus sequence Cys-X2–4-Cys-X15–40-Cys-X2–4-Cys [59][60][61][62] and a helical module [59][62] (Figure 3). The Rossmann fold is composed of six parallel β-strands that are grouped in a central β-sheet surrounded by α-helices. The smaller domain consists of three opposite parallel β-sheets where the tetrahedral zinc ion is bound (Figure 3).
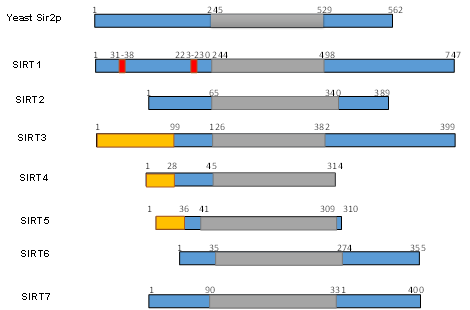
Figure 2. Schematic overview of human SIRTs (SIRT1–7). Human SIRTs are aligned with yeast Sir2p. The conserved large catalytic domain is shown in grey. The nuclear localization sequences shown in red and the mitochondrial targeting sequences are shown in dark yellow. Numbers refer to amino acid residues in the proteins. Adapted from Ref. [9].
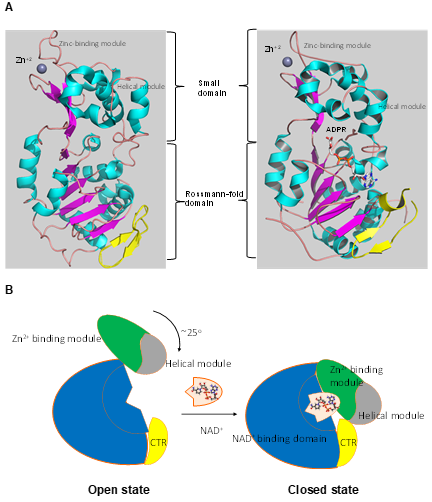
Figure 3. The tertiary structure of SIRT1. (A) The structure in open (left) conformation (SIRT1•CTR complex, PDB code 4IF6) and in closed (right) conformation (SIRT1•CTR•ADPR•Substrate complex, PDB code 4KXQ). The amino acid sequence of SIRT1 is organized into two independent domain. A cavity that is created between the two domains forms the active site. The C-terminal regulatory segment (CTR) is shown in yellow. The CTR binds at the lower edge of the larger NAD+-binding domain, complementing the central parallel β sheet of its Rossmann fold. The figure was created using the program PyMOL (www.pymol.org). (B) Cartoon model of the conformational changes of SIRT1 upon substrate and NAD+ analogue binding. The smaller domain undergoes a rotation with respect to the large domain. A cartoon representation of the apo SIRT1·CTR heterodimer (open state) and the SIRT1·CTR· NAD+·Substrate complex (closed state). Comparison of the open and closed structures revealed that the larger NAD+-binding domain does not undergo any major structural changes. The smaller domain rotates about 25°. The small domain (Zn2+-binding module and the helical module) rotates as a rigid body with only minor changes to the backbone and sidechain conformations. Adapted from Ref. [62].
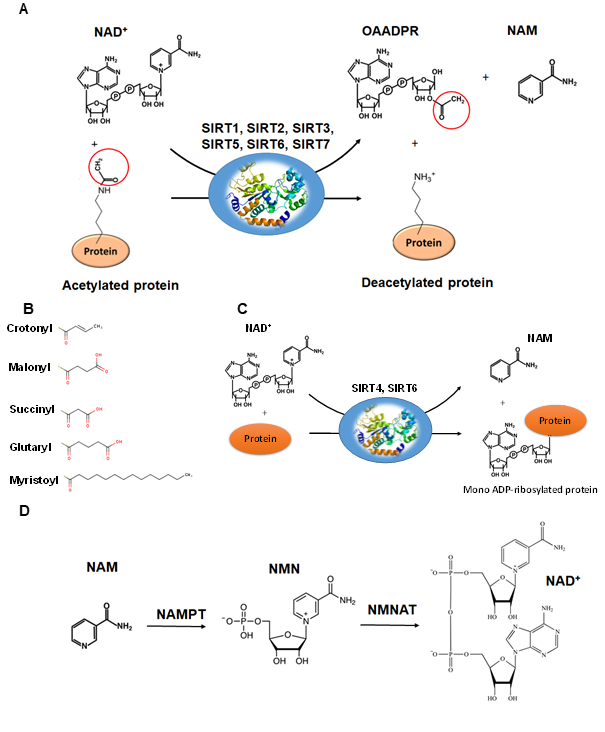
The deacetylation reaction catalyzed by SIRTs is shown in Figure 4A. The reaction includes three substrates: NAD+, water and the acetylated protein. The catalytic cycle involves the formation of the enzyme/NAD+/acetylated substrate ternary complex [63][64][65]. In the deacetylation process, the glycosidic bond between the nicotinamide and ADP-ribose is cleaved and the free nicotinamide (NAM) is released. Then, the acetyl moiety from the substrate is transferred to ADP-ribose to form acetyl-ADP-ribose (2′-O-acetyl-ADP-ribose, AADPR) and the deacetylated protein [63]. The biological role AADPR has not been established so far; however, reasonable experimental results suggest that it acts as a signal transducer . Under normal conditions, AADPR is spontaneously isomerized to 3’-O-acetyl-ADPR.
The active site of SIRTs is formed by an extended “clef” that is responsible for the recognition and binding of the NAD+ cofactor, and is located at the Rossmann fold [63]. The NAD+ binding site can be divided into three sub-regions: (a) the adenine and sugar ribose binding site, (b) the nicotinamide ribose binding site, and (c) the nicotinamide moiety binding site. When the acetylated lysine substrate binds to the enzyme, NAD+ can undergo a conformational change, bringing the nicotinamide group deeper to the c site where it can be cleaved. The active site allows the carbonyl oxygen group of acetylated lysine substrate to come into contact with the anomeric carbon of the nicotinamide riboside of the NAD+. This effective binding accelerates the reaction of acetyl-oxygen with the anomeric carbon, leading to the cleavage of the nicotinamide moiety of NAD+, and the transfer of ADP-ribose to acyl-oxygen, leading to deacetylation.
The deacetylation of lysine residues using as cofactor NAD+ remains the most common reaction that describes the catalytic function of SIRTs more accurately, although there are several examples (Table 1) where they can act on non-acetylated substrates [66][67][68](Figure 4B). For example, it has been reported that succinylated lysine residues in hepatic mitochondria is a target of SIRT5. Another example includes SIRT6, which exhibits demyristoylation activity and has the ability to deacylate long fatty acid aliphatic chains in nuclear factor-κB (NF-κB) factor. SIRT4 acts as an ADP-ribosyltransferase (Figure 4C), whereas SIRT6 exhibits both of these enzymatic activities [69]. SIRT4 also shows lipoamidase activity and SIRT5 displays high desuccinylation activity.
SIRTs are able to recognize many different protein substrates, although they were originally classified exclusively as histone deacetylases [67][68]. For example, SIRT1 has been shown to deacetylate histone H1 on lysine Lys26, H3 on Lys9, Lys14 and Lys56, and H4 on Lys8, Lys12 and Lys16 [67]. The presence of SIRTs in subcellular compartments that do not contain histones prompted the researchers to redefine the range of their protein substrates. Subsequent studies have shown that several other protein substrates are not histones but transcription factors or enzymes that are responsible for cell regulation and adaptation [68].
Figure 4. The reactions catalyzed by SIRTs. (A) The general deacetylase reaction catalyzed by SIRT1,2,3,5,6,7. The natural substrates are NAD+, water and a protein containing acetylated lysine residue. The reaction products include nicotinamide (NAM), the deacetylated protein and the 2’-O-acetyl-ADPR (OAADPR) molecule. (B) Alternative acyl groups that can be deacylated by SIRT3,5,7. (C) The ADP-rebosyltransferase activity catalyzed by SIRT4 and SIRT6. (D) NAM can be used as precursor for the biosynthesis of NAD+ by the enzymes nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyltransferase (NMNAT).
The enzymatic activity of SIRTs can also be affected by post-translational modifications. Phosphorylation is one of the most well-known mechanisms of post-translational regulation of SIRTs [69]. For example, SIRT1 has fifteen phosphorylation sites and several protein kinases, e.g., c-Jun N-terminal kinases (JNKs), casein kinase 2 (CK2), cyclin dependent kinase 1 (CyclinB/Cdk1), dual-specificity tyrosine-phosphorylation-regulated kinases (DYRKs), have been identified with the ability to phosphorylate these sites and thus affect its activity. The phosphorylation and dephosphorylation status of SIRT1 not only influence the catalytic function of the enzyme itself, but also regulate its expression levels through a protease-dependent or independent degradation mechanism [69]. The catalytic function of SIRT2 is also regulated through phosphorylation/dephosphorylation processes. Phosphorylation leads to enzyme activation, whereas dephosphorylation inhibits its activity.
References
- Ren, Z.; He, H.; Zuo, Z.; Xu, Z.; Wei, Z.; Deng, J. The role of different SIRT1-mediated signaling pathways in toxic injury. Cell Mol. Biol Lett. 2019, 24, 1–10.
- Teixeira, C.S.S.; Cerqueira, N.M.F.S.A.; Gomes, P.; Sousa, S.F. A Molecular Perspective on Sirtuin Activity. Int J. Mol. Sci. 2020, 21, 8609.
- Wang, Z.; Guo, W.; Yi, F.; Zhou, T.; Li, X.; Feng, Y.; Guo, Q.; Xu, H.; Song, X.; Cao, L. The Regulatory Effect of SIRT1 on Ex-tracellular Microenvironment Remodeling. Int. J. Biol Sci. 2021, 17, 89–96.
- Soni, S.K.; Basu, P.; Singaravel, M.; Sharma, R.; Pandi-Perumal, S.R.; Cardinali, D.P.; Reiter, R.J. Sirtuins and the circadian clock interplay in cardioprotection: Focus on sirtuin 1. Cell Mol. Life Sci. 2021, in press.
- Ivy, J.M.; Klar, A.J.; Hicks, J.B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell Biol. 1986, 6, 688–702.
- Greiss, S.; Gartner, A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 2009, 28, 407–415.
- Vaquero, A. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 2009, 53, 303–322.
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196.
- Guarente, L. Calorie restriction and sirtuins revisited. Genes Dev. 2013, 27, 2072–2085.
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, doi:10.1038/s41580-020-00313-x.
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Prolla, T.A. Sirt3 mediates reduction of oxidative damage and prevention of agerelated hearing loss under caloric restriction. Cell 2010, 143, 802–812.
- Hwang, J.-W.; Yao, H.; Caito, S.; Sundar, I.K.; Rahman, I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic. Biol. Med. 2013, 61, 95–110.
- Hirschey, M.D.; Shimazu, T.; Goetzman, E.; Jing, E.; Schwer, B.; Lombard, D.B.; Grueter, G.A.; Harris, C.; Biddinger, S.; Ilka-yeva, O.R.; et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010, 464, 121–125.
- Hirschey, M.D.; Shimazu, T.; Jing, E.; Grueter, C.A.; Collins, A.M.; Aouizerat, B.; Verdin, E. SIRT3 deficiency and mitochon-drial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 2011, 44, 177–190.
- Duszka, K.; Gregor, A.; Guillou, H.; König, J.; Wahli, W. Peroxisome Proliferator-Activated Receptors and Caloric Re-striction-Common Pathways Affecting Metabolism, Health, and Longevity. Cells 2020, 9, 1708.
- Yessenkyzy, A.; Saliev, T.; Zhanaliyeva, M.; Masoud, A.R.; Umbayev, B.; Sergazy, S.; Krivykh, E.; Gulyayev, A.; Nurgozhin, T. Polyphenols as Caloric-Restriction Mimetics and Autophagy Inducers in Aging Research. Nutrients 2020, 12, 1344.
- Frye, R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) me-tabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999, 260, 273–279.
- Cen, Y.; Youn, D.Y.; Sauve, A.A. Advances in characterization of human sirtuin isoforms: Chemistries, targets and therapeu-tic applications. Curr. Med. Chem. 2011, 18, 1919–1935.
- Wang, C.H.; Wei, Y.H. Roles of Mitochondrial Sirtuins in Mitochondrial Function, Redx Homeostasis, Insulin Resistance and Type 2 Diabetes. Int. J. Mol. Sci. 2020, 21, 5266.
- Mathias, R.A.; Greco, T.M.; Oberstein, A.; Budayeva, H.G.; Chakrabarti, R.; Rowland, E.A.; Kang, Y.; Shenk, T.; Cristea, I.M. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 2014, 159, 1615–1625.
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013, 18, 920–933.
- Kumar, S.; Lombard, D.B. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit. Rev. Bio-chem. Mol. Biol. 2018, 53, 311–334.
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930.
- Poltronieri, P.; Celetti, A.; Palazzo, L. Mono(ADP-ribosyl)ation Enzymes and NAD+ Metabolism: A Focus on Diseases and Therapeutic Perspectives. Cells 2021, 10, 128. doi: 10.3390/cells10010128.
- Baur, J.A. Biochemical effects of SIRT1 activators. Biochim. Biophys. Acta. 2010, 1804, 1626–1634.
- Sachdeva, V.; Roy, A.; Bharadvaja, N. Current prospects of nutraceuticals: A review. Curr. Pharm. Biotechnol. 2020, 21, 884–896.
- Chalkiadaki, A.; Guarente, L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 2015, 15, 608–624.
- Sánchez-Crisóstomo, I.; Fernández-Martínez, E.; Cariño-Cortés, R.; Betanzos-Cabrera, G.; Bobadilla-Lugo, R.A. Phytosterols and triterpenoids for prevention and treatment of metabolic-related liver diseases and hepatocellular carcinoma. Curr. Pharm. Biotechnol. 2019, 20, 197–214.
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-omics data integration considera-tions and study design for biological systems and disease. Mol. Omics. 2021, doi:10.1039/D0MO00041H.
- Norvell, A.; McMahon, S.B. Rise of the rival. Science 2010, 327, 964–965.
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23.
- Peng, Y.; Li, S.; Landsman, D.; Panchenko, A.R. Histone tails as signaling antennas of chromatin. Curr. Opin. Struct. Biol. 2020, 67, 153–160.
- Villaseñor, R.; Baubec, T. Regulatory mechanisms governing chromatin organization and function. Curr. Opin. Cell Biol. 2020, 70, 10–17.
- Xia, C.; Tao, Y.; Li, M.; Che, T.; Qu, J. Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review). Exp. Ther. Med. 2020, 20, 2923–2940.
- Schreiber SLBernstein, B.E. Signaling network model of chromatin. Cell 2002, 111, 771–778.
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705.
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100.
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793-798.
- Donmez, G.; Guarente, L. Aging and disease: Connections to sirtuins. Aging Cell 2010, 9, 285–290.
- Jiao, F.; Gong, Z. The Beneficial Roles of SIRT1 in Neuroinflammation-Related Diseases. Oxid. Med. Cell Longev. 2020, doi:10.1155/2020/6782872.
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular lo-calizations and functions of human SIRT proteins. Mol. Biol Cell 2005, 16, 4623–4635.
- Tanno, M.; Sakamoto, J.; Miura, T.; Shimamoto, K.; Horio, Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem 2007, 282, 6823–6832.
- Machado, O.R.; Jana, S.; Aleksey, K.; Fleming, O.T. SIRT2 as a Therapeutic Target for Age-Related Disorders. Front. Pharmacol. 2012, 3, 82.
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444.
- Jing, E.; Gesta, S.; Kahn, C.R. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/ deacetylation. Cell Metab. 2007, 6, 105–114.
- Lampson, M.A.; Kapoor, T.M. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 2005, 7, 93–98.
- Ardestani P.M., Liang F. Sub-cellular localization, expression and functions of Sirt6 during the cell cycle in HeLa cells. Nu-cleus 2012, 3, 442–451.
- Haigis, M.; Guarente, L.P.; Farese, R.V.; Weissman, S.; Verdin, E.; Schwer, B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell Biol. 2007, 27, 8807–8814.
- Min, Z.; Gao, J.; Yu, Y. The Roles of Mitochondrial SIRT4 in Cellular Metabolism. Front. Endocrinol. 2019, 9, 783.
- Yang, L.; Miao, S.; Zhang, J.; Wang, P.; Liu, G.; Wang, J. The growing landscape of succinylation links metabolism and heart disease. Epigenomics 2021, doi: 10.2217/epi-2020-0273. Epub ahead of print. PMID: 33605156.
- Yang, L.; Peltier, R.; Zhang, M.; Song, D.; Huang, H.; Chen, G.; Chen, Y.; Zhou, F.; Hao, Q.; Bian, L.; et al. Desuccinyla-tion-Triggered Peptide Self-Assembly: Live Cell Imaging of SIRT5 Activity and Mitochondrial Activity Modulation. J. Am. Chem. Soc. 2020, 142, 18150–18159. doi: 10.1021/jacs.0c08463.
- Yuan, T.; Keijer, J.; Guo, A.H.; Lombard, D.B.; de Boer, V.C.J. An optimized desuccinylase activity assay reveals a difference in desuccinylation activity between proliferative and differentiated cells. Sci. Rep. 2020, 10, 17030.
- Etchegaray, J.P.; Zhong, L.; Mostoslavsky, R. The histone deacetylase SIRT6: At the crossroads between epigenetics, metabo-lism and disease. Curr. Top. Med. Chem. 2013, 13, 2991–3000.
- You, W.; Rotili, D.; Li, T.M.; Kambach, C.; Meleshin, M.; Schutkowski, M.; Chua, K.F.; Mai, A.; Steegborn, C. Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew. Chem. Int. Ed. Engl. 2017, 56, 1007–1011.
- Santos-Barriopedro, I.; Vaquero, A. Complex role of SIRT6 in NF-κB pathway regulation. Mol. Cell Oncol. 2018, 5, e1445942.
- Wu, D.; Li, Y.; Zhu, K.S.; Wang, H.; Zhu, W.G. Advances in Cellular Characterization of the Sirtuin Isoform, SIRT7. Front. Endocrinol. 2018, 9, 652.
- Sacconnay, L.; Carrupt, P.A.; Nurisso, A. Human sirtuins: Structures and flexibility. J. Struct. Biol. 2016, 96, 534–542.
- He, M.; Chiang, H.H.; Luo, H.; Zheng, Z.; Qiao, Q.; Wang, L.; Tan, M.; Ohkubo, R.; Mu, W.C.; Zhao, S.; et al. An Acetylation Switch of the NLRP3 Inflammasome Regulates Aging-Associated Chronic Inflammation and Insulin Resistance. Cell Metab. 2020, 31, 580–591.e5.
- Bordo, D. Structure and evolution of human sirtuins. Curr. Drug Targets 2013, 14, 662–665.
- You, W.; Steegborn, C. Structural Basis for Activation of Human Sirtuin 6 by Fluvastatin. ACS Med. Chem. Lett. 2020, 11, 2285–2289.
- Rumpf, T.; Schiedel, M.; Karaman, B.; Roessler, C.; North, B.J.; Lehotzky, A.; Oláh, J.; Ladwein, K.I.; Schmidtkunz, K.; Gajer, M.; et al. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 2015, 6, 6263.
- Davenport, A.M.; Huber, F.M.; Hoelz, A. Structural and functional analysis of human SIRT1. J. Mol. Biol. 2014, 426, 526–541.
- Hoff, K.G.; Avalos, J.L.; Sens, K.; Wolberger, C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure 2006, 14, 1231–1240.
- Sauve, A.A. Sirtuin chemical mechanisms. Biochim. Biophys. Acta 2010, 1804, 1591–1603.
- Sauve, A.A.; Celic, I.; Avalos, J.; Deng, H.; Boeke, J.D.; Schramm, V.L. Chemistry of gene silencing: The mechanism of NAD+−dependent deacetylation reactions. Biochemistry 2011, 40, 15456–15463.
- Martinez-Redondo, P.; Vaquero, A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer 2013, 4,148–163.
- Bosch-Presegué, L.; Vaquero, A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015, 282, 1745–1767.
- Bheda, P.; Jing, H.; Wolberger, C.; Lin, H. The Substrate Specificity of Sirtuins. Annu. Rev. Biochem. 2016, 85, 405–429.
- Flick, F.; Lüscher, B. Regulation of sirtuin function by posttranslational modifications. Front. Pharmacol. 2012, 3, 29.