| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ewa L. Gajda | + 7235 word(s) | 7235 | 2021-02-19 09:54:03 | | | |
| 2 | Rita Xu | -4072 word(s) | 3163 | 2021-02-25 10:08:51 | | |
Video Upload Options
MicroRNAs (miRNAs, miRs) are small non-coding RNA (ncRNA) molecules capable of regulating post-transcriptional gene expression. Imbalances in the miRNA network have been associated with the development of many pathological conditions and diseases, including cancer. Recently, miRNAs have also been linked to the phenomenon of multidrug resistance (MDR). MiR-7 is one of the extensively studied miRNAs and its role in cancer progression and MDR modulation has been highlighted. MiR-7 is engaged in multiple cellular pathways and acts as a tumor suppressor in the majority of human neoplasia. Its depletion limits the effectiveness of anti-cancer therapies, while its restoration sensitizes cells to the administered drugs.
1. Introduction
The first miRNA (miRNA, miR) was discovered separately by Ambros and Ruvkun in 1993 [1][2]. Nowadays, nearly 2300 miRs have been recognized. Around 50% of miR sequences are accessible in online databases (miRbase; http://www.mirbase.org/; miRBase V22) and multiple software tools allow to predict targets for miRs of choice [3].
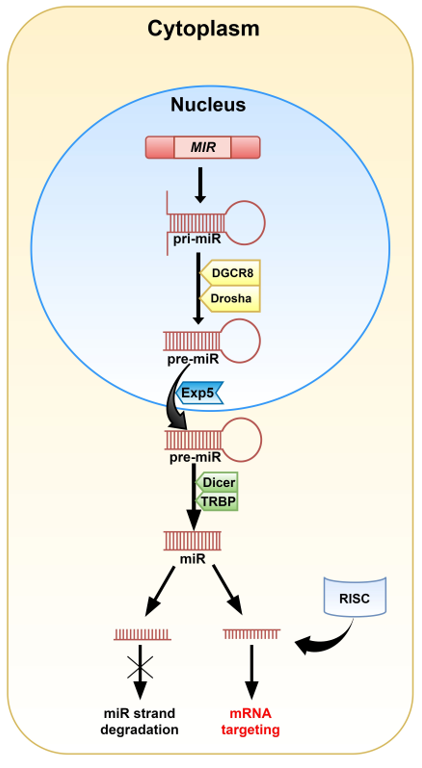
MiRNAs are 20-22 nucleotide-long non-coding RNAs (ncRNAs) that play a crucial role in post-transcriptional regulation of gene expression and their biogenesis is well established. In most cases, miRs are synthesized via the canonical pathway. In this mode, miRs are transcribed individually or as a polycistronic transcript by RNA polymerase II (Pol II), less often by RNA polymerase III (Pol III), in a multistep process. The transcription of miRs is initiated by the formation of primary miRNA (pri-miRNA; Figure 1) in the shape of a hairpin [4][5][6]. The microprocessor complex, containing among others, the DiGeorge critical region 8 (DGCR8) and Drosha enzymes, identifies specific motifs within the sequence of pri-miRNA and releases precursor miRNA (pre-miRNA) by cleaving the stem of the hairpin. This process takes place in the nucleus, but once pre-miRNA is formed, Exportin 5 (Exp5) facilitates its transport to the cytoplasm. The complex made of the Dicer enzyme and TRBP (TAR double-stranded RNA binding protein) captures shuttled pre-miRNA. Dicer presents ribonuclease activity and cuts off the loop of the hairpin and cleaves long double-stranded RNA (dsRNA) into shorter ~20 nucleotide fragments. The generated single-stranded mature miRNA originated from the 5′ and 3′ strands are called 5p and 3p, respectively.
Figure 1. A graphical representation of the canonical pathway of miRNA biogenesis in animals. The MIR gene encodes pri-miRNA. Modification of the pri-miRNA hairpin with two free ends takes place in the cell nucleus. The DGCR8 (DiGeorge critical region 8) and Drosha enzymes cut off the free strands from the hairpin giving pre-miRNA. Then, pre-miRNA is transported into the cytoplasm by Exportin 5 (Exp5), where the Dicer and TRBP (TAR double-stranded RNA binding protein) complex removes the hairpin’s loop and cleaves the molecule into the miRNA duplex. One of the duplex strands, along with the RNA-induced silencing complex (RISC), is involved in mRNA targeting. The second one is degraded.
Biogenesis of miRs via the non-canonical pathways has also been described and is generally divided into two groups: Dicer- and Drosha/DGCR8-independent paths. This mode is often linked with pathological conditions, including cancer [4][7].
The miRNA guide strand anchored to the Argonaute 2 (Ago2) protein targets mRNA during the RNA-induced silencing complex (RISC) loading process [4][5]. Mature miR contains a 2-8 nucleotide-long “seed sequence” that binds to target nucleotides in the 3′ untranslated region of cognate mRNAs, however, binding sites have also been reported in the 5′UTR and coding sequence. The rest of the miR sequence can bind with less complementarity and this allows a miRNA to target multiple mRNAs [4].
The formation of miRNA/Ago2 can harm the production of peptides on various levels, such as: induction of mRNA degradation, affecting proper ribosome assembly and finally, degradation of growing peptides during the process of translation. While the degradation of the mRNA sequence is permanent, the repression of translation can be reversed through detaching miRNA [6]. It has been shown that one microRNA can bind to hundreds of nearly-complementary mRNA sequences and also, one mRNA might be a target for several miRs [8]. MiRs may also cooperate with other ncRNAs including long non-coding RNAs (lncRNAs) [9]. It is assumed that miRs control the expression of nearly 60% of protein-coding genes [10].
MiRNAs are crucial for maintaining cellular development [11][12], differentiation [12][13][14], the cell cycle [15][16][17], proliferation [18][19][20][21], migration [22][23], and apoptosis [24][25]. Apart from their intracellular location, miRNAs are found in biological fluids, such as plasma, saliva, urine, breast milk and might be transferred from one species to another [10][26]. It has been observed that disruption of the miRNA network profile can be linked to several diseases, including cardiovascular diseases, nervous system disorders, and sepsis. Imbalance in the miRNA pool has also been reported in various tumors, such as brain, breast, lung, and colon cancer. MiRNAs may act as either cancer suppressors or as oncogenic factors (oncomiRs). MiR-17-92, miR-21, miR-106, and miR-191 are involved in the development of cancer. Their increased expression has been observed in lung, breast, and gastric cancer, as well as in glioblastoma (GB). On the other hand, depletion of miR-15a, miR-34a, and/or miR-126 suppresses the progression of lung, prostate, and breast tumors [10]. Often, the role of the defined miRNAs is tissue-specific. For example, miR-24 and miR-221/222 are recognized as oncomiRs in breast cancer and glioblastoma, while they act as suppressors in laryngeal or tongue squamous cell carcinoma. Similarly, a dual role of miRNA was observed for miR-155 and miR-125. Lack of miRNA homeostasis in cancer cells promotes enhanced proliferation, angiogenesis, migration, and invasiveness, while blocking apoptosis [27].
It is considered that miRNA can also play a key role in triggering multidrug resistance (MDR) in cancer cells. MDR is a rising therapeutic problem in the treatment of numerous types of tumors as it significantly decreases the effectiveness of anti-cancer drug therapies. Various mechanisms are involved in MDR, including induction of anti-apoptotic machinery or overexpression/activation of several ATP binding cassette (ABC) transporters [28]. Among the 49 ABC proteins, P-glycoprotein (P-gp/ABCB1), breast cancer resistance protein (BCRP/ABCG2), and multidrug resistance-associated protein 1 (MRP1/ABCC1) are the most studied. It has been shown that their high expression correlates with poor prognosis in cancer patients and that a significant portion of cancer-related deaths might be linked with MDR [29][30][31][32][33]. Therefore, there is a need for (i) better understanding of how expression and activity of MDR proteins is managed in tumor cells, (ii) identification of critical genes/proteins/pathways involved in the MDR phenotype of cancer cells, and (iii) elucidation of the role of miRNA in modulation of the MDR phenomenon. Application of antibody- and nano-based vehicles has resulted in major progress in the development of innovative drug delivery systems over the last years [34][35]. Convergence of these strategies with miRNA’s properties might significantly improve therapeutic procedures and effectively impair the progression of diseases, especially in terms of escaping MDR action. The role of miRNA-7 in carcinogenesis and modulation of MDR-encoding genes has been especially highlighted and studied.
2. MiRNA-7
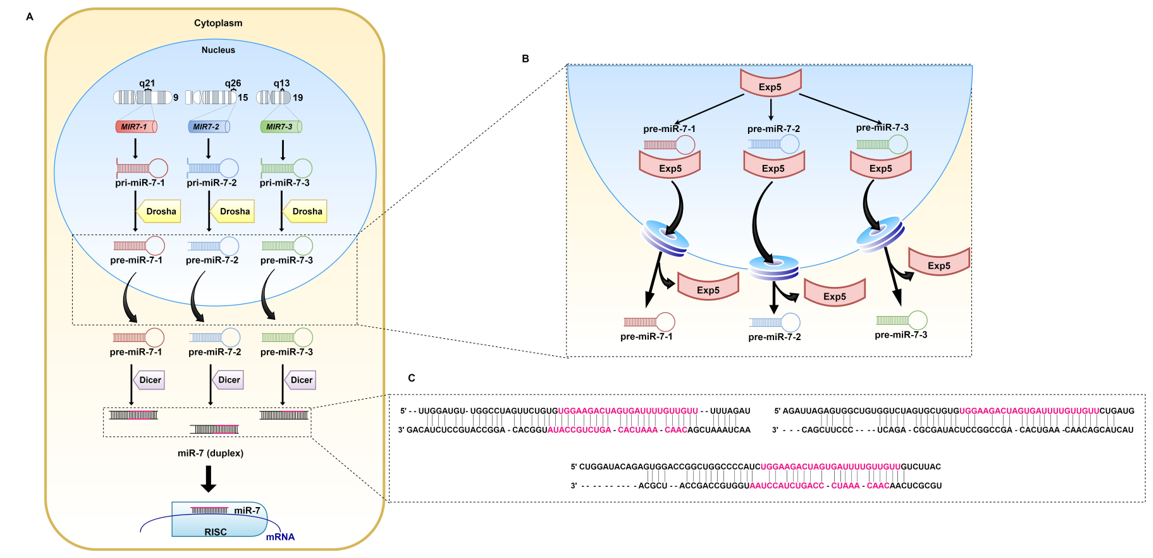
MiRNA-7 (miR-7, hsa-miRNA-7) was first reported in Drosophila melanogaster, nevertheless, the sequence of the guide strand is strongly conserved across different species, which highlights its importance [36]. In humans, miR-7 originates from three precursors: pri-miR-7-1, pri-miR-7-2, and pri-miR-7-3 (Figure 2). They are encoded by the MIR7-1, MIR7-2, and MIR7-3 genes located on three chromosomes: 9q21, 15q26, and 19q13, respectively [37]. Pri-miR-7-1 and pri-miR-7-3 lie within introns of the heterogeneous nuclear ribonucleoprotein K-encoding gene (HNRNPK) and pituitary specific factor 1 gene (PIT1), respectively. The pri-miR-7-2-encoding gene is placed in the intergenic region of chromosome 15 [38][39]. Originally, miR-7 referred to miR-7-5p, since it seemed to be the only mature miRNA from all three precursors that affects cellular pathways. However, other biologically significant miRNAs, miR-7-1-3p and miRNA-7-2-3p, have also been reported [40][41][42][43][44][45]. There are slight changes within the sequence of nucleotides between the miRNAs.
2.1. MiR-7 Biogenesis and Regulation
Mature miR-7 is the product of the canonically transcribed MIR7-1, MIR7-2, and MIR7-3 genes and regulation of transcription proceeds separately for each locus.
Figure 2. Biogenesis of miR-7. (A) miR-7 is the result of transcription of the MIR7-1, MIR7-2, and MIR7-3 genes located on chromosomes 9, 15, and 19, respectively. The generated pri-miRs are transformed into pre-miR-7. Exp5 transports them through the nuclear pores to the cytoplasm (B), where pre-miR-7-1, pre-miR-7-2, and pre-miR-7-3 (C) undergo further modifications. The Dicer complex, cleaves double-stranded RNA (dsRNA) into shorter nucleotide duplexes. Sequences marked in pink are involved in the regulation of mRNA expression.
Apart from regulation of expression of genes encoding for proteins engaged in maturation of miRNA, which applies to all miRNAs, the expression of miR-7 is additionally regulated at the transcriptional level via transcription factors binding to their promoters (Table 1) [36].
It has been shown that transcription of MIR7-1 may be activated by c-Myc [46] and homeobox D10 (HOXD10) [47], whereas hepatocyte nuclear factor 4 alpha (HNF4α) induces MIR7-2 transcription [48]. Another transcriptional factor capable of enhancing miR-7 expression is forkhead box P3 (FOXP3) [49]. An opposite role in miR-7 regulation was revealed for v-Rel avian reticuloendoheliosis viral oncogene homolog A (RELA), which has a binding site in the promotor region of MIR7-1 and MIR7-2. MiR-7 itself also inhibits RELA in a negative feedback manner, by directly binding to the 3′UTR of the RELA transcript [50][51][52]. Ubiquitin-specific protease 18 (Usp18) is another transcription inhibitor of all MIR7. Usp18 decreases the expression of miR-7 host genes, as well as intergenic pri-miR-7-2 [53].
MiR-7 biogenesis is also regulated at the post-transcriptional level. The product of MIR7-1 (pri-miR-7-1) might undergo regulation by the Hu antigen R (HuR) and the Musashi homolog 2 (MSI2) complex. HuR enhances MSI2 binding to the pri-miR-7 conserved terminal loop. This inhibits maturation of pri-miR-7-1 to pre-miR-7-1 [54]. The QKI-5 and QKI-6 proteins restrain miR-7 biogenesis from the MIR7-1 gene. Both proteins directly bind to the pri-miR-7-1 sequence, preventing its further processing and capturing the transcript within the nucleus [55]. The SF2/ASF splicing factor directly connects to pri-miR-7 and supports Drosha in cleavage and maturation [56].
Lastly, mature miR-7 can be modulated by competitive endogenous RNAs (ceRNAs) such as circular RNAs (circRNAs) [57]. They are highly stable and resistant to exonuclease activity due to their covalently closed loop form [58]. The first documented circRNA attenuating miR-7 was derived from the CDR1 gene antisense strand and it is known as ciRS-7 (CDR1as). CiRS-7 is highly expressed in brain and neuronal tissue and contains over 70 seed-matched binding sites for miR-7. It may abate silencing of miR-7 targeted transcripts in brain, non-small cell lung cancer (NSCLC), esophageal squamous cell carcinoma, papillary thyroid cancer, and colorectal cancer [59][60][61][62][63]. CircSNCA is another circular RNA inhibiting miR-7 in brain and neuronal tissue. The treatment-induced muting of circSNCA alters the miR-7 level and induces apoptosis and autophagy in Parkinson’s disease [64]. Gao et al. (2017, 2019), based on microarray analysis, discovered that expression of circ_0006528 is elevated in doxorubicin (DOX, adriamycin)-resistant breast cancer MCF-7 cells and tumor tissues. It was confirmed that circ_0006528 silencing increases expression of miR-7-5p [65][66]. Additionally, Li et al. (2019) indicated that circ-U2AF1 (circ_0061868) also presents direct binding properties to miR-7-5p. U2AF1 encodes for the U2 small nuclear RNA auxiliary factor 1 protein and plays a significant role in RNA splicing as part of the U2 auxiliary complex. The level of circ-U2AF1 is increased in glioma tissues. It was found that downregulation of circ-U2AF1 results in upregulation of miR-7-5p [67]. Until now, researchers have discovered several novel circRNAs working as a miR-7 sponge, such as circ_0000735 in prostate cancer [68], circ-ITCH in osteosarcoma [69], circ-TFCP2L1 in breast cancer [70], and circ_0015756 in hepatocellular carcinoma (HCC) [71].
LncRNAs are an alternative group of ncRNAs that play a significant role in regulating the network of gene expression and miRNAs [72]. Recently, altered expression of lncRNAs was linked with miR-7 silencing in multiple tumors. Zheng et al. (2020) confirmed competitive binding properties of cancer susceptibility 21 (CASC21) lncRNA to miR-7-5p, which results in activation of YAP1 in colorectal cancer [73]. Other lncRNAs targeting miR-7-5p in colorectal cancer are lncRNA RP4, terminal differentiation-induced non-coding RNA (TINCR), and Rhophilin Rho GTPase binding protein 1 antisense RNA 1 (RHPN1-AS1 lcRNA) [74][75][76]. Song et al. (2020) confirmed the effect of RHPN1-AS1 lcRNA in HCC [77]. KCNQ1 overlapping transcript 1 (KCNQ1OT1) lncRNA, which interacts with miR-7-5p, is an example of a lncRNA-based MDR modulator in HCC [78].
It has been reported that lncRNA LINC00115, which is upregulated in triple-negative breast cancer and correlates with poor prognosis, acts as a sponge of miR-7-5p [79]. LINC00115 may also regulate miR-7 expression in lung adenocarcinoma [80]. In renal cell carcinoma, maternally expressed gene 3 (MEG3) lncRNA, which is overexpressed in this type of cancer, targets miR-7 [81]. The activity of MEG3 is frequently decreased in various tumors, such as HCC, cervical, and breast cancer. Another lncRNA found in breast cancer is the Hox transcript antisense intergenic RNA (HOTAIR), which suppresses miR-7 indirectly [82].
FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1) is an oncogenic lncRNA. Thyroid cancer survivors with high expression of FOXD2-AS1 are more prone to relapse. Liu et al. (2019) proved that FOXD2-AS1 has a binding site for miR-7-5p and its downregulation restores a decreased level of miR-7-5p in thyroid carcinoma [83].
Other lncRNAs affecting miR-7-5p include SOX21 antisense RNA 1 (SOX21-AS1) lncRNA in cervical cancer and urothelial cancer associated 1 (UCA1) lncRNA in hypoxia-resistant gastric cancer cells [84][85]. Antisense non-coding RNA in the INK4 locus (ANRIL) is also engaged in tumorigenesis of many tissues. Li et al. (2020) linked the level of miR-7-5p with ANRIL and its role in the progression of T-cell acute lymphoblastic leukemia [86]. LncRNA Cyrano discovered in the brain similarly presents miR-7 sponging capability [87].
Table 1. The list of reported miR-7 regulators in cancer.
|
I. Transcriptional level |
|
|||
|
Transcription factors |
MIR7-1 |
MIR7-2 |
MIR7-3 |
References |
|
c-Myc |
↑ |
— |
— |
[46] |
|
HOXD10 |
↑ |
— |
— |
[47] |
|
HNF4α |
— |
↑ |
— |
[48] |
|
FOXP3 |
↑ |
↑ |
— |
[49] |
|
RELA |
↓ |
↓ |
— |
|
|
Usp18 |
↓ |
↓ |
↓ |
[53] |
|
II. Post-transcriptional level |
|
|||
|
pri-miRNA |
|
|||
|
Factor |
pri-miR-7-1 |
pri-miR-7-2 |
|
|
|
HuR/MSI2 |
↑ |
— |
— |
[54] |
|
QKI-5 |
↓ |
— |
— |
[55] |
|
QKI-6 |
↓ |
— |
— |
[55] |
|
SF2/ASF |
↑ |
— |
— |
[56] |
|
Mature miRNA level |
|
|||
|
ceRNA |
Regulation |
Confirmed in: |
|
|
|
circRNA |
|
|
||
|
ciRS-7 |
↓ |
brain, non-small cell lung cancer, esophageal squamous cell carcinoma, papillary thyroid cancer, colorectal cancer |
||
|
circSNCA |
↓ |
brain |
[64] |
|
|
circ_0006528 |
↓ |
breast cancer |
||
|
circ-U2AF1 |
↓ |
glioma |
[67] |
|
|
circ_0000735 |
↓ |
prostate cancer |
[68] |
|
|
circ-ITCH |
↓ |
osteosarcoma |
[69] |
|
|
circ-TFCP2L1 |
↓ |
breast cancer |
[70] |
|
|
circ_0015756 |
↓ |
hepatocellular carcinoma |
[71] |
|
|
lncRNA |
|
|
||
|
CASC21 |
↓ |
colorectal cancer |
[73] |
|
|
RHPN1-AS1 |
↓ |
colorectal cancer, hepatocellular carcinoma |
||
|
RP4 |
↓ |
colorectal cancer |
[75] |
|
|
TINCR |
↓ |
colorectal cancer |
[76] |
|
|
KCNQ1OT1 |
↓ |
hepatocellular carcinoma |
[78] |
|
|
LINC00115 |
↓ |
triple-negative breast cancer, lung adenocarcinoma |
||
|
MEG3 |
↓ |
renal cell carcinoma |
[81] |
|
|
HOTAIR |
↓ |
breast cancer |
[82] |
|
|
FOXD2-AS1 |
↓ |
thyroid cancer |
[83] |
|
|
SOX21-AS1 |
↓ |
cervical cancer |
[84] |
|
|
UCA1 |
↓ |
gastric cancer |
[85] |
|
|
ANRIL |
↓ |
acute lymphoblastic leukemia |
[86] |
|
|
Cyrano |
↓ |
brain |
[87] |
|
↑, upregulation; ↓, downregulation.
2.2. MiR-7 Expression and Role in Tissues
MiR-7 is particularly critical in tissue of neuroendocrine origin, such as pancreas or brain. It plays an important role in the development and differentiation of those organs [39]. The hypothalamus and pituitary gland are especially enriched in miR-7, in contrast to the cerebellum, cerebral cortex, striatum, or substantia nigra, where its expression is lower. Such an arrangement may be the result of the presence of MIR7-3 in the intron of the PIT1-encoding gene. In the pituitary gland, miR-7 is involved in regulating the secretion of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) via the prostaglandin F2 receptor negative regulator (PTGRF) [39]. Moreover, miR-7 represses the translation of paired box gene 6 (PAX6; important factor in brain and eye organogenesis) by targeting two sites in the 3′UTR [88]. MiR-7 regulates genes involved in repairing neurons [89] and enables synaptic plasticity [90]. Considering the importance of miR-7 in brain tissue, it is not surprising that its downregulation results in the occurrence of pathological conditions like Parkinson’s disease [91] and brain tumors [92][93][94][95]. On the other hand, upregulation of miR-7 is correlated with progression of Alzheimer’s disease [96], schizophrenia [97], and neuroinflammatory processes [98][99].
In the human pancreas, the highest expression of miR-7 occurs between 13 and 18 weeks of gestation. It is correlated with a hormone secretion boost [100]. In the adult pancreas, the islets are characterized by the greatest expression of miR-7 [101], where it regulates proliferation via targeting regenerating islet-derived (Reg) proteins [102]. Moreover, miR-7 is actively engaged in regulation of insulin secretion and its decreased level is correlated with the development of diabetes [103][104]. Lower expression is also observed in pancreatic cancer and might serve as a putative biomarker of disease progression [105][106].
An altered pattern of miR-7 expression and its influence on tumor progression is observed in other types of cancer like breast cancer [51][107], lung cancer [108], melanoma [109], colorectal cancer [110], and hepatocellular carcinoma [111]. Association of miRNA with the formation of multiple tumors may result from a wide range of activity and involvement in primary cellular pathways. MiR-7 regulates proliferation and protects organs against excessive growth. Downregulation of miRNA in tumors leads to uncontrolled proliferation.
Osteosarcoma patients with low levels of miR-7 have poor prognosis [112]. Additionally, cell lines originating from osteosarcoma exhibit enhanced proliferation in comparison to normal osteoblastic cells [112]. Xia et al. (2018) observed that in pancreatic cancer, miRNA-7 causes intensified proliferation through targeting of MAP3K9, hence suppressing NF-κB and MEK/ERK pathways [106]. In pancreatic cancer, increased proliferation is also a consequence of disruption of the EGFR/STAT3 signaling pathway.
Another type of cancer with disruption in the miR-7 status and poor patient outcome is colorectal cancer (CRC). A depletion of miR-7 in CRC also triggers the EGFR pathway [113][114]. Moreover, miR-7 inhibits Krüppel-like factor 4 (KLF4), which acts as an oncogene transcription factor [115]. Another target in CRC is X-ray repair cross complementing 2 (XRCC2), a DNA-repair protein [110]. MiRNA-7 increases proliferation, migration, and angiogenesis through its targets, while decreasing apoptosis in CRC [110][114][115]. In addition, miR-7 blocks metastasis via thyroid receptor interactor protein 6 (TRIP6) [116].
In gastric cancer, miR-7 reduces proliferation and increases apoptosis. However, Lin et al. (2020) linked this function with suppression of Raf-1 [117]. Shi et al. (2014) noticed that expression of both, miR-7 and proteasome activator subunit 3 (REGγ; PSME3), is interfered. REGγ is a direct target for miR-7 and its silencing reduces proliferation and triggers apoptosis [118]. Meanwhile, Wang et al. (2019) investigated the influence of sponging of miR-7 on acceleration of proliferation and migration in triple-negative breast cancer. They associated these findings with serine/threonine-protein kinase PAK 1, which is a direct target for miRNA [70]. Furthermore, miR-7 directly binds to focal adhesion kinase (FAK) triggering downstream effects. In accordance with this, cells showed reduced migration and invasiveness [119]. On the other hand, Yin et al. (2019) found that in glioblastoma, the special AT-rich binding protein 1 (SATB1) is frequently overexpressed. They linked SATB1 with cell migration and invasiveness through its direct impairment by miR-7-5p. The level of miR-7 is also reduced in glioblastoma microvasculature [120]. Restoring its amount prevents extensive proliferation of vascular endothelial cells by targeting Raf-1 [121]. MiR-7 suppresses angiogenesis even in murine xenograft glioblastoma [122]. In melanoma, proliferation and metastasis is inhibited by restoring miR-7 through RELA/NF-κB [109]. In contrast, in non-small cell lung cancer, miR-7 inhibits growth and metastasis via the NOVA alternative splicing regulator 2 (NOVA2) [108] and Bcl-2, a critical regulator of apoptosis [123].
The majority of reports indicate a suppressor role of miR-7 in neoplastic diseases. Restoring the level of miRNA-7 suppresses proliferation and invasiveness and induces apoptosis, reducing malignancy of tumor cells. Since miR-7 is involved in regulation of expression of multiple genes, disrupting its endogenous levels leads to changes in essential signaling pathways. This observation indicates that miR-7 might be a key player in the development of MDR in cancer cells.
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854, doi:10.1016/0092-8674(93)90529-y.
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862, doi:10.1016/0092-8674(93)90530-4.
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364, doi:10.1093/nar/gkz097.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Endocrinol. 2018, 9, 402, doi:10.3389/fendo.2018.00402.
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Rev. Mol. Cell Biol. 2019, 20, 5–20, doi:10.1038/s41580-018-0059-1.
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Genom. 2010, 11, 537–561, doi:10.2174/138920210793175895.
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of MicroRNAs: Many roads to gene regulation. Cells 2019, 8, 1465, doi:10.3390/cells8111465.
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. J. Mol. Sci. 2016, 17, 1712, doi:10.3390/ijms17101712.
- Russo, F.; Fiscon, G.; Conte, F.; Rizzo, M.; Paci, P.; Pellegrini, M. Interplay between long noncoding RNAs and MicroRNAs in In Methods in Molecular Biology; Springer Science and Business Media LLC: Berlin, Germany, 2018; Volume 1819, pp. 75–92.
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276, doi:10.3390/cells9020276.
- Anderson, C.; Catoe, H.; Werner, R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006, 34, 5863–5871, doi:10.1093/nar/gkl743.
- Coffre, M.; Koralov, S.B. miRNAs in B cell development and lymphomagenesis. Trends Mol. Med. 2017, 23, 721–736, doi:10.1016/j.molmed.2017.06.001.
- Chakraborty, M.; Hu, S.; Visness, E.; Del Giudice, M.; De Martino, A.; Bosia, C.; Sharp, P.A.; Garg, S. MicroRNAs organize intrinsic variation into stem cell states. Natl. Acad. Sci. USA 2020, 117, 6942–6950, doi:10.1073/pnas.1920695117.
- Foshay, K.M.; Gallicano, G.I. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Biol. 2009, 326, 431–443, doi:10.1016/j.ydbio.2008.11.016.
- Liu, Q.; Fu, H.; Sun, F.; Zhang, H.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008, 36, 5391–5404, doi:10.1093/nar/gkn522.
- Ivanovska, I.; Ball, A.S.; Diaz, R.L.; Magnus, J.F.; Kibukawa, M.; Schelter, J.M.; Kobayashi, S.V.; Lim, L.; Burchard, J.; Jackson, A.L.; et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Cell. Biol. 2008, 28, 2167–2174, doi:10.1128/mcb.01977-07.
- Borden, A.; Kurian, J.; Nickoloff, E.; Yang, Y.; Troupes, C.D.; Ibetti, J.; Lucchese, A.M.; Gao, E.; Mohsin, S.; Koch, W.J.; et al. Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Res. 2019, 125, 14–25, doi:10.1161/circresaha.118.314223.
- Wu, J.; Qian, J.; Li, C.; Kwok, L.; Cheng, F.; Liu, P.; Perdomo, C.; Kotton, D.; Vaziri, C.; Anderlind, C.; et al. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle 2010, 9, 1809–1818, doi:10.4161/cc.9.9.11535.
- Liu, C.; Teng, Z.-Q.; Santistevan, N.J.; Szulwach, K.E.; Guo, W.; Jin, P.; Zhao, X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 2010, 6, 433–444, doi:10.1016/j.stem.2010.02.017.
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; Oday, E.M.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR MicroRNA recognition elements. Cell 2009, 35, 610–625, doi:10.1016/j.molcel.2009.08.020.
- Singh, A.P.; Hung, Y.-H.; Shanahan, M.T.; Kanke, M.; Bonfini, A.; Dame, M.K.; Biraud, M.; Peck, B.C.; Oyesola, O.O.; Freund, J.M.; et al. Enteroendocrine progenitor cell–enriched miR-7 regulates intestinal epithelial proliferation in an Xiap-dependent manner. Mol. Gastroenterol. Hepatol. 2020, 9, 447–464, doi:10.1016/j.jcmgh.2019.11.001.
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Cell 2010, 39, 133–144, doi:10.1016/j.molcel.2010.06.010.
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523, doi:10.1038/emboj.2012.183.
- Xu, C.; Lu, Y.; Pan, Z.; Chu, W.; Luo, X.; Lin, H.; Xiao, J.; Shan, H.; Wang, Z.; Yang, B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. Cell Sci. 2007, 120, 3045–3052, doi:10.1242/jcs.010728.
- Buscaglia, L.E.B.; Li, Y. Apoptosis and the target genes of microRNA-21. J. Cancer 2011, 30, 371–380, doi:10.5732/cjc.011.10132.
- Lukasik, A.; Zielenkiewicz, P. Plant MicroRNAs—Novel players in natural medicine? J. Mol. Sci. 2016, 18, 9, doi:10.3390/ijms18010009.
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56, doi:10.4331/wjbc.v8.i1.45.
- Tsuruo, T.; Naito, M.; Tomida, A.; Fujita, N.; Mashima, T.; Sakamoto, H.; Haga, N. Molecular targeting therapy of cancer: Drug resistance, apoptosis and survival signal. Cancer Sci. 2003, 94, 15–21, doi:10.1111/j.1349-7006.2003.tb01345.x.
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792, doi:10.3390/cancers6031769.
- Sun, Y.-L.; Patel, A.; Kumar, P.; Chen, Z.-S. Role of ABC transporters in cancer chemotherapy. J. Cancer 2012, 31, 51–57, doi:10.5732/cjc.011.10466.
- Wu, S.; Fu, L. Tyrosine kinase inhibitors enhanced the efficacy of conventional chemotherapeutic agent in multidrug resistant cancer cells. Cancer 2018, 17, 25, doi:10.1186/s12943-018-0775-3.
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. Pathol. 2005, 205, 275–292, doi:10.1002/path.1706.
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Pharm. Bull. 2017, 7, 339–348, doi:10.15171/apb.2017.041.
- Majidinia, M.; Mirza‐Aghazadeh‐Attari, M.; Rahimi, M.; Mihanfar, A.; Karimian, A.; Safa, A.; Yousefi, B. Overcoming multidrug resistance in cancer: Recent progress in nanotechnology and new horizons. IUBMB Life 2020, 72, 855–871, doi:10.1002/iub.2215.
- Dallavalle, S.; Dobričić, V.; Lazzarato, L.; Gazzano, E.; Machuqueiro, M.; Pajeva, I.; Tsakovska, I.; Zidar, N.; Fruttero, R. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist. Updates 2020, 50, 100682, doi:10.1016/j.drup.2020.100682.
- Horsham, J.L.; Ganda, C.; Kalinowski, F.C.; Brown, R.A.; Epis, M.R.; Leedman, P.J. MicroRNA-7: A miRNA with expanding roles in development and disease. J. Biochem. Cell Biol. 2015, 69, 215–224, doi:10.1016/j.biocel.2015.11.001.
- Zhao, J.; Tao, Y.; Zhou, Y.; Qin, N.; Chen, C.; Tian, D.; Xu, L. MicroRNA-7: A promising new target in cancer therapy. Cancer Cell Int. 2015, 15, 103, doi:10.1186/s12935-015-0259-0.
- Kalinowski, F.C.; Brown, R.A.; Ganda, C.; Giles, K.M.; Epis, M.R.; Horsham, J.; Leedman, P.J. microRNA-7: A tumor suppressor miRNA with therapeutic potential. J. Biochem. Cell Biol. 2014, 54, 312–317, doi:10.1016/j.biocel.2014.05.040.
- Zhao, J.; Zhou, Y.; Guo, M.; Yue, D.; Chen, C.; Liang, G.; Xu, L. MicroRNA-7: Expression and function in brain physiological and pathological processes. Cell Biosci. 2020, 10, 77, doi:10.1186/s13578-020-00436-w.
- Horsham, J.L.; Kalinowski, F.C.; Epis, M.R.; Ganda, C.; Brown, R.A.M.; Leedman, P.J. Clinical potential of microRNA-7 in cancer. Clin. Med. 2015, 4, 1668–1687, doi:10.3390/jcm4091668.
- Mohorianu, I.; Fowler, E.K.; Dalmay, T.; Chapman, T. Control of seminal fluid protein expression via regulatory hubs in Drosophila melanogaster. R. Soc. B Biol. Sci. 2018; 285, 20181681.
- Liu, K.; Feng, F.; Yang, Y.; Duan, J.; Liu, H.; Yang, J.; Wu, M.; Liu, C.; Chang, Y. High-throughput screening identified miR-7-2-3p and miR-29c-3p as metastasis suppressors in gallbladder carcinoma. Gastroenterol. 2019, 55, 51–66, doi:10.1007/s00535-019-01627-0.
- Wu, H.; Wei, Y.; Pan, S. Down‐regulation and clinical significance of miR‐7‐2‐3p in papillary thyroid carcinoma with multiple detecting methods. IET Syst. Biol. 2019, 13, 225–233, doi:10.1049/iet-syb.2019.0025.
- Pallarès-Albanell, J.; Zomeño-Abellán, M.T.; Escaramís, G.; Pantano, L.; Soriano, A.; Segura, M.F.; Martí, E. A high-throughput screening identifies MicroRNA inhibitors that influence neuronal maintenance and/or response to oxidative stress. Ther. Nucleic Acids 2019, 17, 374–387, doi:10.1016/j.omtn.2019.06.007.
- Chakrabarti, M.; Ray, S.K. Anti-tumor activities of luteolin and silibinin in glioblastoma cells: Overexpression of miR-7-1-3p augmented luteolin and silibinin to inhibit autophagy and induce apoptosis in glioblastoma in vivo. Apoptosis 2015, 21, 312–328, doi:10.1007/s10495-015-1198-x.
- Chou, Y.-T.; Lin, H.-H.; Lien, Y.-C.; Wang, Y.-H.; Hong, C.-F.; Kao, Y.-R.; Lin, S.-C.; Chang, Y.-C.; Lin, S.-Y.; Chen, S.-J.; et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010, 70, 8822–8831, doi:10.1158/0008-5472.can-10-0638.
- Reddy, S.D.N.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008, 68, 8195–8200, doi:10.1158/0008-5472.can-08-2103.
- Ning, B.-F.; Ding, J.; Liu, J.; Yin, C.; Xu, W.-P.; Cong, W.-M.; Zhang, Q.; Chen, F.; Han, T.; Deng, X.; et al. Hepatocyte nuclear factor 4α-nuclear factor-κB feedback circuit modulates liver cancer progression. Hepatology 2014, 60, 1607–1619, doi:10.1002/hep.27177.
- McInnes, N.; Sadlon, T.J.; Brown, C.Y.; Pederson, S.; Beyer, M.; Schultze, J.L.; McColl, S.; Goodall, G.J.; Barry, S.C. FOXP3 and FOXP3-regulated microRNAs suppress SATB1 in breast cancer cells. Oncogene 2011, 31, 1045–1054, doi:10.1038/onc.2011.293.
- Choi, D.C.; Chae, Y.-J.; Kabaria, S.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Mouradian, M.M.; Junn, E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. Neurosci. 2014, 34, 12725–12737, doi:10.1523/jneurosci.0985-14.2014.
- Li, M.; Pan, M.; Wang, J.; You, C.; Zhao, F.; Zheng, D.; Guo, M.; Xu, H.; Wu, D.; Wang, L.; et al. miR-7 reduces breast cancer stem cell metastasis via inhibiting RELA to decrease ESAM expression. Ther. Oncolytics 2020, 18, 70–82, doi:10.1016/j.omto.2020.06.002.
- Zhao, X.-D.; Lu, Y.-Y.; Guo, H.; Xie, H.-H.; He, L.-J.; Shen, G.-F.; Zhou, J.-F.; Li, T.; Hu, S.-J.; Zhou, L.; et al. MicroRNA-7/NF-κB signaling regulatory feedback circuit regulates gastric carcinogenesis. Cell Biol. 2015, 210, 613–627, doi:10.1083/jcb.201501073.
- Duex, J.E.; Comeau, L.; Sorkin, A.; Purow, B.; Kefas, B. Usp18 regulates epidermal growth factor (EGF) receptor expression and cancer cell survival via MicroRNA-7. Biol. Chem. 2011, 286, 25377–25386, doi:10.1074/jbc.m111.222760.
- Choudhury, N.R.; Alves, F.D.L.; De Andrés-Aguayo, L.; Graf, T.; Cáceres, J.F.; Rappsilber, J.; Michlewski, G. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013, 27, 24–38, doi:10.1101/gad.199190.112.
- Wang, Y.; Vogel, G.; Yu, Z.; Richard, S. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of MicroRNA 7 in glial cells. Cell. Biol. 2013, 33, 1233–1243, doi:10.1128/mcb.01604-12.
- Wu, H.; Sun, S.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.R.; Zhu, J. A splicing-independent function of SF2/ASF in MicroRNA processing. Cell 2010, 38, 67–77, doi:10.1016/j.molcel.2010.02.021.
- Mitra, A.; Pfeifer, K.; Park, K.-S. Circular RNAs and competing endogenous RNA (ceRNA) networks. Cancer Res. 2018, 7, S624–S628, doi:10.21037/tcr.2018.05.12.
- Zhao, W.; Dong, M.; Pan, J.; Wang, Y.; Zhou, J.; Ma, J.; Liu, S. Circular RNAs: A novel target among non‑coding RNAs with potential roles in malignant tumors (review). Med. Rep. 2019, 20, 3463–3474, doi:10.3892/mmr.2019.10637.
- Su, C.; Han, Y.; Zhang, H.; Li, Y.; Yi, L.; Wang, X.; Zhou, S.; Yu, D.; Song, X.; Xiao, N.; et al. CiRS‐7 targeting miR‐7 modulates the progression of non‐small cell lung cancer in a manner dependent on NF‐κB signalling. Cell. Mol. Med. 2018, 22, 3097–3107, doi:10.1111/jcmm.13587.
- Huang, H.; Wei, L.; Qin, T.; Yang, N.; Li, Z.; Xu, Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol. Ther. 2019, 20, 73–80, doi:10.1080/15384047.2018.1507254.
- Han, J.-Y.; Guo, S.; Wei, N.; Xue, R.; Li, W.; Dong, G.; Li, J.; Tian, X.; Chen, C.; Qiu, S.; et al. ciRS-7 Promotes the proliferation and migration of papillary thyroid cancer by negatively regulating the miR-7/epidermal growth factor receptor axis. BioMed Res. Int. 2020, 2020, 1–14, doi:10.1155/2020/9875636.
- Tang, W.; Ji, M.; He, G.; Yang, L.; Niu, Z.; Jian, M.; Wei, Y.; Ren, L.; Xu, J. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther. 2017, 10, 2045–2056, doi:10.2147/ott.s131597.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388, doi:10.1038/nature11993.
- Sang, Q.; Liu, X.; Wang, L.; Qi, L.; Sun, W.; Wang, W.; Sun, Y.; Zhang, H. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 2018, 10, 1281–1293, doi:10.18632/aging.101466.
- Gao, D.; Qi, X.; Zhang, X.; Fang, K.; Guo, Z.; Li, L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Carcinog. 2019, 58, 554–564, doi:10.1002/mc.22950.
- Gao, D.; Zhang, X.; Liu, B.; Meng, D.; Fang, K.; Guo, Z.; Li, L. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017, 9, 1175–1188, doi:10.2217/epi-2017-0055.
- Li, G.; Huang, M.; Cai, Y.; Yang, Y.; Sun, X.; Ke, Y. Circ‐U2AF1 promotes human glioma via derepressing neuro‐oncological ventral antigen 2 by sponging hsa‐miR‐7‐5p. Cell. Physiol. 2019, 234, 9144–9155, doi:10.1002/jcp.27591.
- Gao, Y.; Liu, J.; Huan, J.; Che, F. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020, 20, 1–13, doi:10.1186/s12935-020-01421-6.
- Li, H.; Lan, M.; Liao, X.; Tang, Z.; Yang, C. Circular RNA cir-ITCH promotes osteosarcoma migration and invasion through cir-ITCH/miR-7/EGFR pathway. Cancer Res. Treat. 2020, 19, 1533033819898728, doi:10.1177/1533033819898728.
- Wang, Q.; Li, Z.; Hu, Y.; Zheng, W.; Tang, W.; Zhai, C.; Gu, Z.; Tao, J.; Wang, H. Circ-TFCP2L1 promotes the proliferation and migration of triple negative breast cancer through sponging miR-7 by inhibiting PAK1. Mammary Gland. Biol. Neoplasia 2019, 24, 323–331, doi:10.1007/s10911-019-09440-4.
- Liu, L.; Yang, X.; Li, N.-F.; Lin, L.; Luo, H. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle 2019, 18, 2939–2953, doi:10.1080/15384101.2019.1664223.
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Cell Biol. 2019, 21, 542–551, doi:10.1038/s41556-019-0311-8.
- Zheng, Y.; Nie, P.; Xu, S. Long noncoding RNA CASC21 exerts an oncogenic role in colorectal cancer through regulating miR-7-5p/YAP1 axis. Pharmacother. 2020, 121, 109628, doi:10.1016/j.biopha.2019.109628.
- Zheng, W.; Li, H.; Zhang, H.; Zhang, C.; Zhu, Z.; Liang, H.; Zhou, Y. Long noncoding RNA RHPN1-AS1 promotes colorectal cancer progression via targeting miR-7-5p/OGT axis. Cancer Cell Int. 2020, 20, 1–12, doi:10.1186/s12935-020-1110-9.
- Liu, M.-L.; Zhang, Q.; Yuan, X.; Jin, L.; Wang, L.-L.; Fang, T.-T.; Wang, W.-B. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World J. Gastroenterol. 2018, 24, 1004–1012, doi:10.3748/wjg.v24.i9.1004.
- Eckhart, L.; Lachner, J.; Tschachler, E.; Rice, R.H. TINCR is not a non‐coding RNA but encodes a protein component of cornified epidermal keratinocytes. Dermatol. 2020, 29, 376–379, doi:10.1111/exd.14083.
- Song, X.-Z.; Ren, X.-N.; Xu, X.-J.; Ruan, X.-X.; Wang, Y.-L.; Yao, T.-T. LncRNA RHPN1-AS1 promotes cell proliferation, migration and invasion through targeting miR-7-5p and activating PI3K/AKT/mTOR pathway in hepatocellular carcinoma. Cancer Res. Treat. 2020, 19, 1533033820957023, doi:10.1177/1533033820957023.
- Hu, H.; Yang, L.; Li, L.; Zeng, C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ ABCC1 axis. Biophys. Res. Commun. 2018, 503, 2400–2406, doi:10.1016/j.bbrc.2018.06.168.
- Yuan, C.; Luo, X.; Duan, S.; Guo, L. Long noncoding RNA LINC00115 promotes breast cancer metastasis by inhibiting miR‐7. FEBS Open Bio 2020, 10, 1230–1237, doi:10.1002/2211-5463.12842.
- Li, D.S.; Ainiwaer, J.L. Sheyhiding, I.; Zhang, Z.; Zhang, L.W. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Rev. Pharmacol. Sci. 2016, 20, 2285–2295.
- He, H.; Dai, J.; Zhuo, R.; Zhao, J.; Wang, H.; Sun, F.; Zhu, Y.; Xu, D. Study on the mechanism behind lncRNA MEG3 affecting clear cell renal cell carcinoma by regulating miR‐7/RASL11B signaling. Cell. Physiol. 2018, 233, 9503–9515, doi:10.1002/jcp.26849.
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by LincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868, doi:10.1002/stem.1795.
- Liu, X.; Fu, Q.; Li, S.; Liang, N.; Li, F.; Li, C.; Sui, C.; Dionigi, G.; Sun, H. LncRNA FOXD2-AS1 functions as a competing endogenous RNA to regulate TERT expression by sponging miR-7-5p in thyroid cancer. Endocrinol. 2019, 10, 207, doi:10.3389/fendo.2019.00207.
- Zhang, X.; Zhao, X.; Li, Y.; Zhou, Y.; Zhang, Z. Long noncoding RNA SOX21‐AS1 promotes cervical cancer progression by competitively sponging miR‐7/VDAC1. Cell. Physiol. 2019, 234, 17494–17504, doi:10.1002/jcp.28371.
- Yang, Z.; Shi, X.; Li, C.; Wang, X.; Hou, K.; Li, Z.; Zhang, X.; Fan, Y.; Qu, X.; Che, X.; et al. Long non-coding RNA UCA1 upregulation promotes the migration of hypoxia-resistant gastric cancer cells through the miR-7-5p/EGFR axis. Cell Res. 2018, 368, 194–201, doi:10.1016/j.yexcr.2018.04.030.
- Li, G.; Gao, L.; Zhao, J.; Liu, D.; Li, H.; Hu, M. LncRNA ANRIL/miR-7-5p/TCF4 axis contributes to the progression of T cell acute lymphoblastic leukemia. Cancer Cell Int. 2020, 20, 1–12, doi:10.1186/s12935-020-01376-8.
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 2018, 174, 350–362.e17, doi:10.1016/j.cell.2018.05.022.
- Needhamsen, M.; White, R.B.; Giles, K.M.; Dunlop, S.A.; Thomas, M.G. Regulation of human PAX6 expression by miR-7. Bioinform. Online 2014, 10, 107–113, doi:10.4137/ebo.s13739.
- Fan, Z.; Lu, M.; Qiao, C.; Zhou, Y.; Ding, J.-H.; Hu, G. MicroRNA-7 enhances subventricular zone neurogenesis by inhibiting NLRP3/caspase-1 axis in adult neural stem cells. Neurobiol. 2016, 53, 7057–7069, doi:10.1007/s12035-015-9620-5.
- Hu, G.; Niu, F.; Liao, K.; Periyasamy, P.; Sil, S.; Liu, J.; Dravid, S.M.; Buch, S. HIV-1 tat-induced astrocytic extracellular vesicle miR-7 impairs synaptic architecture. Neuroimmune Pharmacol. 2020, 15, 538–553, doi:10.1007/s11481-019-09869-8.
- Li, S.; Lv, X.; Zhai, K.; Xu, R.; Zhang, Y.; Zhao, S.; Qin, X.; Yin, L.; Lou, J. MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax and Sirt2. J. Transl. Res. 2016, 8, 993–1004.
- Gajda, E.; Godlewska, M.; Mariak, Z.; Nazaruk, E.; Gawel, D. Combinatory treatment with miR-7-5p and drug-loaded cubosomes effectively impairs cancer cells. J. Mol. Sci. 2020, 21, 5039, doi:10.3390/ijms21145039.
- Visani, M.; De Biase, D.; Marucci, G.; Cerasoli, S.; Nigrisoli, E.; Reggiani, M.L.B.; Albani, F.; Baruzzi, A.; Pession, A.; The PERNO Study Group. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I–III. Oncol. 2013, 8, 417–430, doi:10.1016/j.molonc.2013.12.010.
- Kumar, V.; Kumar, V.; Chaudhary, A.K.; Coulter, D.W.; McGuire, T.; Mahato, R.I. Impact of miRNA-mRNA profiling and their correlation on medulloblastoma tumorigenesis. Ther. Nucleic Acids 2018, 12, 490–503, doi:10.1016/j.omtn.2018.06.004.
- Chen, H.; Shalom-Feuerstein, R.; Riley, J.; Zhang, S.-D.; Tucci, P.; Agostini, M.; Aberdam, D.; Knight, R.A.; Genchi, G.; Nicotera, P.; et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biophys. Res. Commun. 2010, 394, 921–927, doi:10.1016/j.bbrc.2010.03.076.
- Madadi, S.; Schwarzenbach, H.; Saidijam, M.; Mahjub, R.; Soleimani, M. Potential microRNA-related targets in clearance pathways of amyloid-β: Novel therapeutic approach for the treatment of Alzheimer’s disease. Cell Biosci. 2019, 9, 91, doi:10.1186/s13578-019-0354-3.
- Zhang, J.; Sun, X.-Y.; Zhang, L.-Y. MicroRNA-7/Shank3 axis involved in schizophrenia pathogenesis. Clin. Neurosci. 2015, 22, 1254–1257, doi:10.1016/j.jocn.2015.01.031.
- Yue, D.; Zhao, J.; Chen, H.; Guo, M.; Chen, C.; Zhou, Y.; Xu, L. MicroRNA-7, synergizes with RORα, negatively controls the pathology of brain tissue inflammation. Neuroinflammation 2020, 17, 28, doi:10.1186/s12974-020-1710-2.
- Zhang, X.-D.; Fan, Q.-Y.; Qiu, Z.; Chen, S. MiR-7 alleviates secondary inflammatory response of microglia caused by cerebral hemorrhage through inhibiting TLR4 expression. Rev. Med. Pharmacol. Sci. 2018, 22, 5597–5604.
- Correa-Medina, M.; Bravo-Egana, V.; Rosero, S.; Ricordi, C.; Edlund, H.; Diez, J.; Pastori, R.L. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr. Patterns 2009, 9, 193–199, doi:10.1016/j.gep.2008.12.003.
- Wang, Y.; Liu, J.; Liu, C.; Naji, A.; Stoffers, D.A. MicroRNA-7 regulates the mTOR Pathway and proliferation in adult pancreatic β-cells. Diabetes 2012, 62, 887–895, doi:10.2337/db12-0451.
- Downing, S.; Zhang, F.; Chen, Z.; Tzanakakis, E.S. MicroRNA-7 directly targets Reg1 in pancreatic cells. J. Physiol. Physiol. 2019, 317, C366–C374, doi:10.1152/ajpcell.00013.2019.
- Nieto, M.; Hevia, P.; Garcia, E.; Klein, D.; Alvarez-Cubela, S.; Bravo-Egana, V.; Rosero, S.; Molano, R.D.; Vargas, N.; Ricordi, C.; et al. Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplant. 2012, 21, 1761–1774, doi:10.3727/096368911x612521.
- Matarese, A.; Gambardella, J.; Lombardi, A.; Wang, X.; Santulli, G. miR-7 regulates GLP-1-mediated insulin release by targeting β-arrestin 1. Cells 2020, 9, 1621, doi:10.3390/cells9071621.
- Ye, Z.-Q.; Zou, C.-L.; Chen, H.-B.; Jiang, M.-J.; Mei, Z.; Gu, D.-N. MicroRNA-7 as a potential biomarker for prognosis in pancreatic cancer. Markers 2020, 2020, 1–13, doi:10.1155/2020/2782101.
- Xia, J.; Cao, T.; Ma, C.; Shi, Y.; Sun, Y.; Wang, Z.P.; Ma, J. miR-7 suppresses tumor progression by directly targeting MAP3K9 in pancreatic cancer. Ther. Nucleic Acids 2018, 13, 121–132, doi:10.1016/j.omtn.2018.08.012.
- Moazzeni, H.; Najafi, A.; Khani, M. Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Cell. Probes 2017, 34, 45–52, doi:10.1016/j.mcp.2017.05.005.
- Xiao, H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Mol. Biol. Lett. 2019, 24, 1–13, doi:10.1186/s11658-019-0188-3.
- Giles, K.M.; Brown, R.A.; Ganda, C.; Podgorny, M.J.; Candy, P.A.; Wintle, L.C.; Richardson, K.L.; Kalinowski, F.C.; Stuart, L.M.; Epis, M.R.; et al. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-κB. Oncotarget 2016, 7, 31663–31680, doi:10.18632/oncotarget.9421.
- Xu, K.; Song, X.; Chen, Z.; Qin, C. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014, 7, 325–332, doi:10.2147/ott.s59364.
- Wu, W.; Liu, S.; Liang, Y.; Zhou, Z.; Liu, X. MiR-7 inhibits progression of hepatocarcinoma by targeting KLF-4 and promises a novel diagnostic biomarker. Cancer Cell Int. 2017, 17, 31, doi:10.1186/s12935-017-0386-x.
- Liu, S.; Zhou, C.; Zhu, C.; Song, Q.; Wen, M.; Liu, Y.; An, H. Low-expression of miR-7 promotes cell proliferation and exhibits prognostic value in osteosarcoma patients. J. Clin. Exp. Pathol. 2017, 10, 9035–9041.
- Suto, T.; Yokobori, T.; Yajima, R.; Morita, H.; Fujii, T.; Yamaguchi, S.; Altan, B.; Tsutsumi, S.; Asao, T.; Kuwano, H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2014, 36, 338–345, doi:10.1093/carcin/bgu242.
- Fan, X.; Liu, M.; Tang, H.; Leng, D.; Hu, S.; Lu, R.; Wan, W.; Yuan, S. MicroRNA-7 exerts antiangiogenic effect on colorectal cancer via ERK signaling. Surg. Res. 2019, 240, 48–59, doi:10.1016/j.jss.2019.02.035.
- Dong, M.; Xie, Y.; Xu, Y. miR‑7‑5p regulates the proliferation and migration of colorectal cancer cells by negatively regulating the expression of Krüppel‑like factor 4. Lett. 2019, 17, 3241–3246, doi:10.3892/ol.2019.10001.
- Ling, Y.; Cao, C.; Li, S.; Qiu, M.; Shen, G.; Chen, Z.; Yao, F.; Chen, W. TRIP6, as a target of miR-7, regulates the proliferation and metastasis of colorectal cancer cells. Biophys. Res. Commun. 2019, 514, 231–238, doi:10.1016/j.bbrc.2019.04.092.
- Lin, J.; Liu, Z.; Liao, S.; Li, E.; Wu, X.; Zeng, W. Elevated microRNA-7 inhibits proliferation and tumor angiogenesis and promotes apoptosis of gastric cancer cells via repression of Raf-1. Cell Cycle 2020, 19, 2496–2508, doi:10.1080/15384101.2020.1807670.
- Shi, Y.; Luo, X.; Li, P.; Tan, J.; Wang, X.; Xiang, T.; Ren, G. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett. 2015, 358, 27–36, doi:10.1016/j.canlet.2014.12.014.
- Kong, X.; Li, G.; Yuan, Y.; He, Y.; Wu, X.; Zhang, W.; Wu, Z.; Chen, T.; Wu, W.; Lobie, P.E.; et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE 2012, 7, e41523, doi:10.1371/journal.pone.0041523.
- Yin, C.; Kong, W.; Jiang, J.; Xu, H.; Zhao, W. miR‑7‑5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Lett. 2018, 17, 1819–1825, doi:10.3892/ol.2018.9777.
- Liu, Z.; Liu, Y.; Li, L.; Xu, Z.; Bi, B.; Wang, Y.; Li, J.Y. MiR-7-5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumor Biol. 2014, 35, 10177–10184, doi:10.1007/s13277-014-2318-x.
- Babae, N.; Bourajjaj, M.; Liu, Y.; Van Beijnum, J.R.; Cerisoli, F.; Scaria, P.V.; Verheul, M.; Van Berkel, M.P.; Pieters, E.H.E.; Van Haastert, R.J.; et al. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 2014, 5, 6687–6700, doi:10.18632/oncotarget.2235.
- Xiong, S.; Zheng, Y.; Jiang, P.; Liu, R.; Liu, X.; Chu, Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. J. Biol. Sci. 2011, 7, 805–814, doi:10.7150/ijbs.7.805.