
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rita B. Santos | + 1427 word(s) | 1427 | 2021-02-19 07:56:02 | | | |
| 2 | Lily Guo | Meta information modification | 1427 | 2021-02-25 06:56:20 | | |
Video Upload Options
Proteases are an integral part of plant defense systems, with several hubs of action, from pathogen recognition and priming to the activation of plant hypersensitive response. Within this wide group of proteolytic enzymes, aspartic proteases have been implicated in several plant development functions and are gaining more prominence due to their involvement in plant–pathogen interactions.
1. The Past and the Present of Aspartic Proteases
Aspartic proteases (APs) were first discovered in animals during the nineteenth century. In 1836, Theodor Schwann described pepsin, which he identified during the study of animal gastric juices [1]. Later, in 1875, a pepsin-like proteinase was described in the pitcher plant (Nepenthes) after treatment of the plants’ digestive juice with sulfuric acid [2]. Almost a century after the discovery of the first AP, in 1930, the purification and crystallization of swine pepsin by John Northrop provided a substantial evidence that proteases were proteins[3]. In the following years, other proteases were crystallized and studied, including chymotrypsin, trypsin, and pepsinogen[4]. The conversion of pepsinogen to the active form of pepsin is an autocatalytic process that occurs at a low pH (1.5–5). Based on these findings, in 1962, the first step was taken towards the study of acidic proteinases[5]. In 1970, the discovery of pepstatin[6], a powerful inhibitor of aspartic proteases, encouraged its use as an immobilized compound for affinity purification of these proteases [7][8]. A major breakthrough occurred in 1972, when the complete amino acid sequence of the pig pepsin was uncovered[9]. Later, in the 1980s, the current terminology of the aspartic (or aspartyl) proteases was established, resulting from the observation that the carboxyl groups belonging to aspartate residues were involved in the catalytic process [10].
Although most studies about APs were performed in mammals, yeast, and fungi, some work has been developed in plants[11]. Plant APs were purified from the seeds of many organisms, such as Oryza sativa [12], Cucurbita maxima[13], Cucumis sativus [13], Triticum aestivum [14], and Hordeum vulgare[15], as well as from Lycopersicon esculentum leaves [16]. In 1991, the DNA sequence of the first plant aspartic protease, from barley (Hordeurn vulgare), was sequenced [17]. In the following years, APs from other organisms, including one from Arabidopsis thaliana, were isolated, providing more information about plant APs’ structure[7].
According to the MEROPS database (http://www.merops.ac.uk), aspartic proteases (EC 3.4.23) are grouped into 16 families, based on similarities of the amino acid sequences of the catalytic site. These families are clustered into five different clans that reflect a common evolutionary origin and similar tertiary structure [18]. Plant aspartic proteases are distributed among 12 of the 16 families: A1, A2, A3, A9, A11, A28, and A32 of clan AA; families A22 and A24 of clan AD; family A8 and A31 of clan AC and AE, respectively; and family A36 which has not yet been assigned to a clan[19]. A majority of plant APs belong to A1 family[20].
In 2004, with the completion of Arabidopsis genome, new perspectives have risen regarding plant APs’ diversity [21]. The first plant aspartic protease gene family to be described belonged to Arabidopsis with 51 known genes[22]. In the following years, plant APs have been found in increasing numbers[23] with 96 OsAP genes in rice (Oryza sativa)[24], 50 VvAP genes in grapevine (Vitis vinifera)[25], and 67 PtAP genes identified in black cottonwood (Populus trichocarpa)[26].
So far, it is known that plant APs are involved in several cell mechanisms, from developmental processes[27][28][29] to abiotic[30][32][31] and biotic stress responses[33][34][35]. Major milestones concerning aspartic proteases’ history and relation to pathogen resistance are presented in Figure 1.
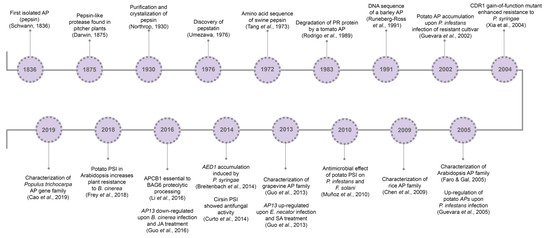
Figure 1. Aspartic proteases historical breakthroughs. AP, aspartic protease; CDR1, constitutive disease resistance 1; PSI, plant-specific insert; AP13, aspartic protease 13; SA, salicylic acid; AED1, apoplastic enhanced disease susceptibility 1; APCB1, aspartyl protease cleaving bcl-2 associated athanogene; BAG6, BCL-2 associated athanogene protein 6; SA, salicylic acid; JA, jasmonic acid; MeJA, methyl jasmonate.
2. The Features of Plant Aspartic Proteases
2.1. Structure and Classification
In the past years, with the study of phytepsin (AP from barley seeds) and cardosin (AP from the flowers of Cynara cardunculus), relevant information has been generated about plant aspartic proteases [22][36][37]. Plant APs, mostly belonging to family A1, are generally active at acid pH (pH 2–6), are specifically inhibited by pepstatin A, and comprise two aspartic acid residues essential for the catalytic activity[20][38]. The catalytic motifs of plant aspartic proteases from A1 family are usually Asp-Thr-Gly (DTG) or Asp-Ser-Gly (DSG)[20]. Although the general structure of the plant APs has similarities to that of mammals and microorganisms, plant APs contain a plant-specific insert (PSI) in the C-terminal region [7].
Most of the knowledge about A1 family plant APs comes from the study of typical APs, such as phytepsin and cardosin A and B [20]. Typical APs possess a signal peptide, a prosegment, and a PSI, and the catalytic site is composed by hydrophobic-hydrophobic-DTG-Ser-Ser residues (Figure 2). Exceptions to the structure of typical aspartic proteases were already described, as in the case of nucellin [39], in chloroplast nucleoid DNA-binding protein (CND41) [40], and in the constitutive disease resistance 1 (CDR1) protease[33]. These structural exceptions gave rise to three different categories, depending on the putative domain organization and active site sequence motifs: typical, nucellin-like, and atypical aspartic proteases [22]. Atypical and nucellin-like APs have distinct features on primary structure organization that differ from typical APs. The nucellin-like APs lack the prosegment and the PSI and comprise proteins similar to nucellin[20] with a characteristic sequence of residues: acidic-hydrophobic-DTG-serine-acidic residues around the catalytic site[22]. Atypical APs have intermediate features between typical and nucellin-like, and the active site is composed by hydrophobic-hydrophobic-DTG-Ser-acidic residues [20]. Both atypical and nucellin-like APs have a cysteine-rich region designated nepenthesin-type AP (NAP) specific insertion[38].
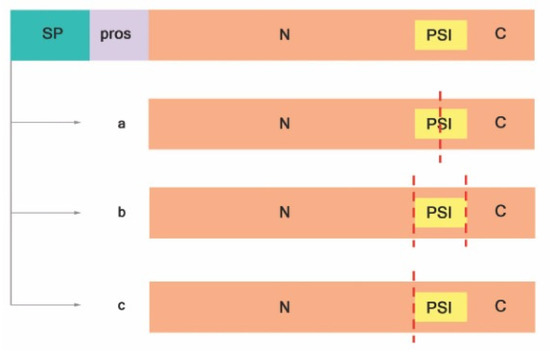
Figure 2. Proteolytic activation of typical APs adapted from Reference [41]. (a) PSI is digested at the midsection, (b) PSI is entirely removed, and (c) PSI and C-terminal are removed. Signal peptide (SP); prosegment (pros); plant-specific insert (PSI); N-terminal domain (N); C-terminal domain (C); red dashed lines indicate cleavage sites.
Detailed information on structure organization of plant aspartic proteases has been extensively reviewed in Reference [38].
2.2. Activation of Aspartic Proteases and Their Subcellular Localization
Proteolytic cleavage is crucial for active proteases. It starts with the removal of the signal sequence upon translocation to the ER lumen resulting in proproteins (zymogens). Usually, processing zymogens of typical plant APs involves the removal of the prosegment and partial or total deletion of PSI in an autocatalytic manner at the low pH of the vacuole[20][41]. Cheung and colleagues have proposed that, after proteolytic cleavage and activation, typical plant APs are either heterodimeric, where the PSI is partially digested (Figure 2a) [42][43][44] or entirely removed (Figure 2b; Reference [45]), or monomeric, without PSI (Figure 2c), as was observed in sweet potato SPAP1 [46].
There is evidence that two monomeric APs from potato tuber and leaves, StAsp1 [47] and StAsp3 [48], respectively, have the PSI in their mature form. However, the proteolytic mechanisms behind that process are still unknown [49][50]. In contrast, two atypical aspartic proteases, CDR1 and its rice homolog, have shown activity without the removal of the putative prosegment[51][52]. More studies have to be conducted to fully understand the inactivation mechanisms of plant APs. Soares and colleagues have recently proposed that the inactive form of APs occurs because the active site is blocked by the prosegment alone or by the prosegment together with the mature N-terminal and the flap. In contrast, precursors of cardosin A and B are active before undergoing the proteolytic process that removes prosegment, hence probably do not share the inactivation mechanism described above[20].
Considering the APs’ subcellular location, these proteases are found in various cellular compartments. Typical APs are mostly found in vacuoles, such as in the case of APs from barley [53], castor bean (Ricinus communis) [54], and Arabidopsis[55]. To a less extent, typical APs are also located in the extracellular space, such as in the case of tomato (Solanum lycopersicum)[16] and tobacco (Nicotiana tabacum) APs [56]. Atypical APs are widely distributed in the cell: Arabidopsis PCS1, ASPG1, and ASPR1 are located in the endoplasmic reticulum (ER) [29][30][57]; UNDEAD AP in mitochondria [58]; and CND41 and NANA in the chloroplast[27][40]. Rice OsAP65 is located in pre-vacuolar compartments [28]; nepenthesins and Arabidopsis AED1 and CDR1 are distributed in the extracellular space [33][59][60]. Arabidopsis A36 and A39 APs were found to be located in the plasma membrane as anchored proteins[61].
Plant APs are involved in many biological functions, particularly in developmental processes, such as chloroplast homeostasis and protein turnover[27][40], as well as in programmed cell death (PCD) and cell survival[62]. Developmentally controlled plant cell death is initiated through hormonal signaling, which in turn leads to the accumulation of reactive oxygen species (ROS) and transcriptional activation of PCD-related genes, such as proteases and nucleases. PCD can have different outcomes, such as senescence, the death of cells no longer required, or the creation of tissues that assume structural storage functions[63].
References
- Schwann, T. Ueber das Wesen des Verdauungsprocesses. Ann. Phys. Chem. 1836, 114, 358–364, doi:10.1002/andp.18361140612.
- Darwin, C. Insectivorous Plants; Murray, J., Ed.; John Murray: London, UK, 1875.
- Northrop, J.H. Crystalline pepsin: I. Isolation and tests of purity. J. Gen. Physiol. 1930, 13, 739–766, doi:10.1085/jgp.13.6.739.
- Northrop, J.H. Crystalline Enzymes. The Chemistry of Pepsin, Trypsin, and Bacteriophage; Columbia University Press: New York, NY, USA, 1939.
- Herriott, R.M. Pepsinogen and pepsin. J. Gen. Physiol. 1962, 45, 57–76, doi:10.1085/jgp.45.4.57.
- Umezawa, H. Enzyme Inhibitors of Microbial Origin; University of Tokyo Press: Tokyo, Japan, 1972.
- Mutlu, A.; Gal, S. Plant aspartic proteinases: Enzymes on the way to a function. Physiol. Plant. 1999, 105, 569–576, doi:10.1034/j.1399-3054.1999.105324.x.
- Fruton, J.S. A history of pepsin and related enzymes. Q. Rev. Biol. 2002, 77, 127–147, doi:10.1086/340729.
- Tang, J.; Sepulveda, P.; Marciniszyn, J.; Chen, K.C.; Huang, W.Y.; Tao, N.; Liu, D.; Lanier, J.P. Amino acid sequence of por-cine pepsin. Proc. Natl. Acad. Sci. USA. 1973, 70, 3437–3439, doi:10.1073/pnas.70.12.3437.
- Fruton, J.S. Aspartyl proteinases. In Hydrolytic Enzymes; Brocklehurst, K., Neuberger, A., Eds.; Elsevier: Amsterdam, The Netherlands, 1987; p. 423; ISBN 9780080860756.
- Szecsi, P.B. The aspartic proteases. Scand. J. Clin. Lab. Investig. 1992, 52, 2–22, doi:10.3109/00365519209104650.
- Dol, E.; Shibata, D.; Matoba, T.; Yoneza, W.A.D. Characterization of pepstatin-sensitive acid protease in resting rice seeds. Agric. Biol. Chem. 1980, 44, 741–747, doi:10.1080/00021369.1980.10864028.
- Polanowski, A.; Wilusz, T.; Kolaczkowska, M.K.; Wieczorek, M.; Wilimowska-Pelc, A. Purification and characterization of aspartic proteinases from Cucumis sativus and Cucurbita maxima seeds. In Aspartic Proteinases and Their Inhibitors; Kostka, V., Ed.; Springer, Boston, MA, USA, 1985; pp. 49–52.
- Dunaevsky, Y.E.; Sarbakanova, S.T.; Belozersky, M.A. Wheat seed carboxypeptidase and joint action on gliadin of proteases from dry and germinating seeds. J. Exp. Bot. 1989, 40, 1323–1329.
- Sarkkinen, P.; Kalkkinen, N.; Tilgmann, C.; Siuro, J.; Kervinen, J.; Mikola, L. Aspartic proteinase from barley grains is re-lated to mammalian lysosomal cathepsin D. Planta 1992, 186, 317–323, doi:10.1007/BF00195311.
- Rodrigo, I.; Vera, P.; Conejero, V. Degradation of tomato pathogenesis‐related proteins by an endogenous 37‐kDa aspartyl endoproteinase. Eur. J. Biochem. 1989, 184, 663–669, doi:10.1111/j.1432-1033.1989.tb15064.x.
- Runeberg-Roos, P.; Tormakangas, K.; Ostman, A. Primary structure of a barley-grain aspartic proteinase. A plant aspartic proteinase resembling mammalian cathepsin D. Eur. J. Biochem. 1991, 202, 1021–1027, doi:10.1111/j.1432-1033.1991.tb16465.x.
- Paparelli, E.; Gonzali, S.; Parlanti, S.; Novi, G.; Giorgi, F.M.; Licausi, F.; Kosmacz, M.; Feil, R.; Lunn, J.E.; Brust, H.; et al. Mi-sexpression of a chloroplast aspartyl protease leads to severe growth defects and alters carbohydrate metabolism in Ara-bidopsis. Plant Physiol. 2012, 160, 1237–1250, doi:10.1104/pp.112.204016.
- Huang, J.; Zhao, X.; Cheng, K.; Jiang, Y.; Ouyang, Y.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. J. Exp. Bot. 2013, 64, 3351–3360, doi:10.1093/jxb/ert173.
- Soares, A.; Niedermaier, S.; Faro, R.; Loos, A.; Manadas, B.; Faro, C.; Huesgen, P.F.; Cheung, A.Y.; Simões, I. An atypical aspartic protease modulates lateral root development in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 2157–2171, doi:10.1093/jxb/erz059.
- Yao, X.; Xiong, W.; Ye, T.; Wu, Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Ara-bidopsis. J. Exp. Bot. 2012, 63, 2579–2593, doi:10.1093/jxb/err433.
- Sebastián, D.I.; Fernando, F.D.; Raúl, D.G.; Gabriela, G.M. Overexpression of Arabidopsis aspartic protease APA1 gene confers drought tolerance. Plant Sci. 2020, 292, 110406, doi:10.1016/j.plantsci.2020.110406.
- Guo, R.; Zhao, J.; Wang, X.X.; Guo, C.; Li, Z.; Wang, Y.; Wang, X.X. Constitutive expression of a grape aspartic protease gene in transgenic Arabidopsis confers osmotic stress tolerance. Plant Cell Tissue Organ Cult. 2015, 121, 275–287, doi:10.1007/s11240-014-0699-6.
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease func-tions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988, doi:10.1038/sj.emboj.7600086.
- Alam, M.M.; Nakamura, H.; Ichikawa, H.; Miyao, A.; Hirochika, H.; Kobayashi, K.; Yamaoka, N.; Nishiguchi, M. Response of an aspartic protease gene OsAP77 to fungal, bacterial and viral infections in rice. Rice 2014, 7, 9, doi:10.1186/s12284-014-0009-2.
- Guo, R.; Tu, M.; Wang, X.; Zhao, J.; Wan, R.; Li, Z.; Wang, Y.; Wang, X. Ectopic expression of a grape aspartic protease gene, AP13, in Arabidopsis thaliana improves resistance to powdery mildew but increases susceptibility to Botrytis cinerea. Plant Sci. 2016, 248, 17–27, doi:10.1016/j.plantsci.2016.04.006.
- Faro, C.; Ramalho-Santos, M.; Veríssimo, P.; Pissarra, J.; Frazão, C.; Costa, J.; Lin, X.-L.; Tang, J.; Pires, E. Structural and functional aspects of cardosins. In Advances in Experimental Medicine and Biology; Springer LLC: New York, NY, USA, 1998; Volume 436, pp. 423–433.
- Kervinen, J.; Tobin, G.J.; Costa, J.; Waugh, D.S.; Wlodawer, A.; Zdanov, A. Crystal structure of plant aspartic proteinase prophytepsin: Inactivation and vacuolar targeting. EMBO J. 1999, 18, 3947–3955, doi:10.1093/emboj/18.14.3947.
- Soares, A.; Ribeiro Carlton, S.M.; Simões, I. Atypical and nucellin-like aspartic proteases: Emerging players in plant devel-opmental processes and stress responses. J. Exp. Bot. 2019, 70, 2059–2076, doi:10.1093/jxb/erz034.
- Chen, F.; Foolad, M.R. Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 1997, 35, 821–831, doi:10.1023/A:1005833207707.
- Nakano, T.; Murakami, S.; Shoji, T.; Yoshida, S.; Yamada, Y.; Sato, F. A novel protein with DNA binding activity from to-bacco chloroplast nucleoids. Plant Cell 1997, 9, 1673–1682, doi:10.1105/tpc.9.9.1673.
- Cheung, L.K.Y.; Dupuis, J.H.; Dee, D.R.; Bryksa, B.C.; Yada, R.Y. Roles of Plant-Specific Inserts in Plant Defense. Trends Plant Sci. 2020, doi:10.1016/j.tplants.2020.02.009.
- Park, H.; Kusakabe, I.; Sakakibara, Y.; Kobayashi, H. Autoproteolytic processing of aspartic proteinase from sunflower seeds. Biosci. Biotechnol. Biochem. 2001, 65, 702–705, doi:10.1271/bbb.65.702.
- Castanheira, P.; Samyn, B.; Sergeant, K.; Clemente, J.C.; Dunn, B.M.; Pires, E.; Van Beeumen, J.; Faro, C. Activation, proteo-lytic processing, and peptide specificity of recombinant cardosin A. J. Biol. Chem. 2005, 280, 13047–13054, doi:10.1074/jbc.M412076200.
- Feijoo-Siota, L.; Rama, J.L.R.; Sánchez-Pérez, A.; Villa, T.G. Expression, activation and processing of a novel plant milk-clotting aspartic protease in Pichia pastoris. J. Biotechnol. 2018, 268, 28–39, doi:10.1016/j.jbiotec.2018.01.006.
- Ramalho-Santos, M.; Verissimo, P.; Cortes, L.; Samyn, B.; Van Beeumen, J.; Pires, E.; Faro, C. Identification and proteolytic processing of procardosin A. Eur. J. Biochem. 1998, 255, 133–138, doi:10.1046/j.1432-1327.1998.2550133.x.
- Chen, H.-J.; Huang, Y.-H.; Huang, G.-J.; Huang, S.-S.; Chow, T.-J.; Lin, Y.-H. Sweet potato SPAP1 is a typical aspartic protease and participates in ethephon-mediated leaf senescence. J. Plant Physiol. 2015, 180, 1–17, doi:10.1016/j.jplph.2015.03.009.
- Guevara, M.G.; Oliva, C.R.; Machinandiarena, M.; Daleo, G.R. Purification and properties of an aspartic protease from pota-to tuber that is inhibited by a basic chitinase. Physiol. Plant. 1999, 106, 164–169, doi:10.1034/j.1399-3054.1999.106203.x.
- Guevara, M.G.; Daleo, G.R.; Oliva, C.R. Purification and characterization of an aspartic protease from potato leaves. Physiol. Plant. 2001, 112, 321–326, doi:10.1034/j.1399-3054.2001.1120304.x.
- Guevara, M.G.; Almeida, C.; Mendieta, J.R.; Faro, C.J.; Veríssimo, P.; Pires, E.V.; Daleo, G.R. Molecular cloning of a potato leaf cDNA encoding an aspartic protease (StAsp) and its expression after P. infestans infection. Plant Physiol. Biochem. 2005, 43, 882–889, doi:10.1016/j.plaphy.2005.07.004.
- Frey, M.E.; D’Ippolito, S.; Pepe, A.; Daleo, G.R.; Guevara, M.G. Transgenic expression of plant-specific insert of potato aspar-tic proteases (StAP-PSI) confers enhanced resistance to Botrytis cinerea in Arabidopsis thaliana. Phytochemistry 2018, 149, 1–11, doi:10.1016/j.phytochem.2018.02.004.
- Simões, I.; Faro, R.; Bur, D.; Faro, C. Characterization of recombinant CDR1, an Arabidopsis aspartic proteinase involved in disease resistance. J. Biol. Chem. 2007, 282, 31358–31365, doi:10.1074/jbc.M702477200.
- Prasad, B.D.; Creissen, G.; Lamb, C.; Chattoo, B.B. Heterologous expression and characterization of recombinant OsCDR1, a rice aspartic proteinase involved in disease resistance. Protein Expr. Purif. 2010, 72, 169–174, doi:10.1016/j.pep.2010.03.018.
- Runeberg-Roos, P.; Kervinen, J.; Kovaleva, V.; Raikhel, N.V.; Gal, S. The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro. Plant Physiol. 1994, 105, 321–329, doi:10.1104/pp.105.1.321.
- Hiraiwa, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants. Eur. J. Biochem. 1997, 246, 133–141, doi:10.1111/j.1432-1033.1997.00133.x.
- Mutlu, A.; Chen, X.; Reddy, S.M.; Gal, S. The aspartic proteinase is expressed in Arabidopsis thaliana seeds and localized in the protein bodies. Seed Sci. Res. 1999, 9, 75–84, doi:10.1017/s0960258599000082.
- Rodrigo, I.; Vera, P.; Van Loon, L.C.; Conejero, V. Degradation of tobacco pathogenesis-related proteins: Evidence for con-served mechanisms of degradation of pathogenesis-related proteins in plants. Plant Physiol. 1991, 95, 616–622, doi:10.1104/pp.95.2.616.
- Ge, X.; Dietrich, C.; Matsuno, M.; Li, G.; Berg, H.; Xia, Y. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep. 2005, 6, 282–288, doi:10.1038/sj.embor.7400357.
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224, doi:10.1105/tpc.110.082651.
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jordá, L.; Maldonado-Alconada, A.M.; Sarioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contrasting roles of the apoplastic aspartyl protease Apoplastic, Enhanced Disease Suscepti-bility1-Dependent1 and Legume Lectin-Like Protein1 in Arabidopsis systemic acquired resistance. Plant Physiol. 2014, 165, 791–809, doi:10.1104/pp.114.239665.
- Athauda, S.B.P.; Matsumoto, K.; Rajapakshe, S.; Kuribayashi, M.; Kojima, M.; Kubomura-Yoshida, N.; Iwamatsu, A.; Shibata, C.; Inoue, H.; Takahashi, K. Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochem. J. 2004, 381, 295–306, doi:10.1042/BJ20031575.
- Gao, H.; Li, R.; Guo, Y. Arabidopsis aspartic proteases A36 and A39 play roles in plant reproduction. Plant Signal. Behav. 2017, 12, e1304343, doi:10.1080/15592324.2017.1304343.
- Stael, S.; Van Breusegem, F.; Gevaert, K.; Nowack, M.K. Plant Proteases and Programmed Cell Death; Oxford University Press: Oxford, UK, 2019; Volume 70, pp. 1991–1995.
- Huysmans, M.; Lema, A.S.; Coll, N.S.; Nowack, M.K. Dying two deaths—Programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 2017.
- Rawlings, N.D.; Barrett, A.J. MEROPS: The peptidase database. Nucleic Acids Res. 1999, 27, 325–331, doi:10.1093/nar/27.1.325.
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic en-zymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632, doi:10.1093/nar/gkx1134.
- Simões, I.; Faro, C. Structure and function of plant aspartic proteinases. Eur. J. Biochem. 2004, 271, 2067–2075.
- Beers, E.P.; Jones, A.M.; Dickerman, A.W. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phy-tochemistry 2004, 65, 43–58, doi:10.1016/j.phytochem.2003.09.005.
- Faro, C.; Gal, S. Aspartic Proteinase Content of the Arabidopsis Genome. Curr. Protein Pept. Sci. 2005, 6, 493–500, doi:10.2174/138920305774933268.
- Dunn, B.M. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002, 102, 4431–4458, doi:10.1021/cr010167q.
- Chen, J.; Ouyang, Y.; Wang, L.; Xie, W.; Zhang, Q. Aspartic proteases gene family in rice: Gene structure and expression, predicted protein features and phylogenetic relation. Gene 2009, 442, 108–118, doi:10.1016/j.gene.2009.04.021.
- Guo, R.; Xu, X.; Carole, B.; Li, X.; Gao, M.; Zheng, Y.; Wang, X. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC Genom. 2013, 14, 554, doi:10.1186/1471-2164-14-554.
- Cao, S.; Guo, M.; Wang, C.; Xu, W.; Shi, T.; Tong, G.; Zhen, C.; Cheng, H.; Yang, C.; Elsheery, N.I.; et al. Genome-wide char-acterization of aspartic protease (AP) gene family in Populus trichocarpa and identification of the potential PtAPs involved in wood formation. BMC Plant Biol. 2019, 19, 276, doi:10.1186/s12870-019-1865-0.




