| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alan Zanardi | + 2128 word(s) | 2128 | 2021-02-07 07:56:52 | | | |
| 2 | Rita Xu | -881 word(s) | 1247 | 2021-02-23 11:09:56 | | |
Video Upload Options
Neurodegenerative disorders can induce modifications of several proteins; one of which is ceruloplasmin (Cp), a ferroxidase enzyme found modified in the cerebrospinal fluid (CSF) of neurodegenerative diseases patients.
1. Introduction
The occurrence of aberrant post-translational modifications in brain proteins is a feature reported in different neurodegenerative diseases, including Amyotrophic Lateral Sclerosis (ALS), Alzheimer’s (AD) and Parkinson’s (PD) disease [1][2][3][4]. These modifications generally involve proteins directly related to the pathology, such as superoxide dismutase, TDP-43 protein, Aβ peptide, Tau protein and α-synuclein [1][2][3][4] but are not limited to them.
Post-translational modifications of ceruloplasmin (Cp), a ferroxidase enzyme present inter alia in the brain, have been observed in the cerebrospinal fluid (CSF) of patients suffering from neurodegeneration [5][6][7][8]. The Cp modifications are usually associated with loss of enzymatic activity and are promoted by the pathological environment [5][6][7][8]. One of the Cp modifications, found in the CSF from both AD and PD patients, is deamidation [9][10], a spontaneous process connected to protein aging [11]. Protein deamidation occurs on two amino acid residues, namely asparagine and glutamine, and generally results in loss of protein function [11]. In the case of Cp, the deamidation involves asparagine residues at the level of two asparagine-glycine-arginine (Asn-Gly-Arg, NGR) sites of the protein sequence whose, upon deamidation, gain binding activity to integrins [9][10][12]. Integrins are a family of receptors that mediated cell-extracellular matrix adhesion [13]. The binding of modified Cp to integrins triggers in vitro intracellular signaling on epithelial cells, altering the cell physiology with the induction of detrimental effects [9][12]. However, despite the presence of NGR motifs deamidation in the Cp from neurodegenerative diseases patients’ CSF, in vivo cellular targets and possible pathological effects are still unexplored topics.
2. Protein Deamidation, Unwanted Phenomenon or Regulated Process?
Asparagine (Asn, N) deamidation is a spontaneous reaction in which the amide functional group of Asn is removed, and the amino acid is converted to either an aspartate (Asp, D) or an isoaspartate (isoAsp, isoD) residue [11]. Glutamine (Gln, Q) can also be subjected to deamidation, but at lower rate compared to Asn [14]. It has been demonstrated that deamidation is a post-translational modification that spontaneously occurs in vivo, under physiological conditions [15]. The process is autocatalytic and nonenzymatic, and it depends on the protein primary, secondary, and tertiary structure [16]. The rate of deamidation is also determined by the cellular environment, for example by temperature, ionic composition and strength, and pH changes [11].
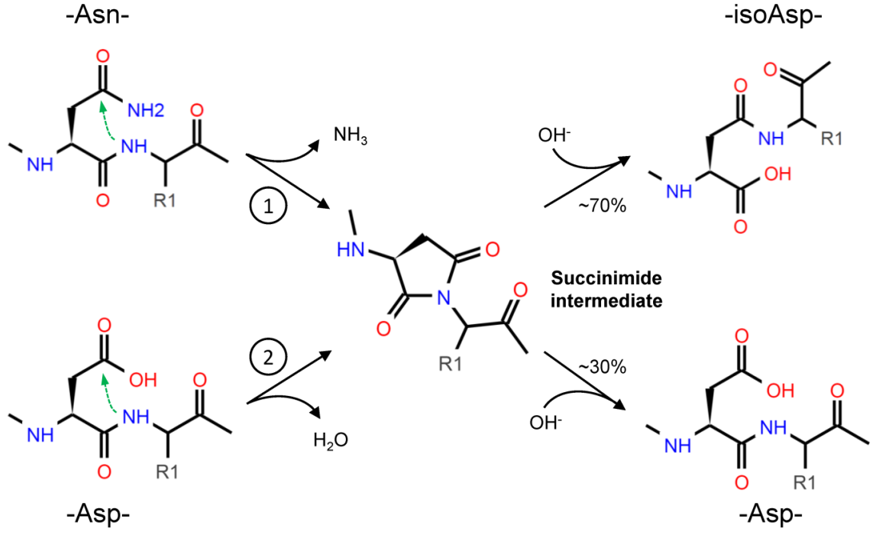
Through deamidation, Asn forms a cyclic succinimide intermediate, which is then hydrolyzed to either Asp or isoAsp; due to the asymmetry of the succinimide, isoAsp is the primary product (Figure 1). The formation of isoAsp can also arise from dehydration of Asp, but at slower rate than Asn residues [17]. Although in a smaller proportion, the racemization of succinimide can even result in D-Asp and D-isoAsp [17][18]. Depending on the combination of the conditions mentioned above (sequence, structure, temperature, etc.), the spontaneous deamidation of specific residues can take different times to happen: for some residues, it will occur very quickly, while for some others it will never occur [11][19].
Figure 1. Asparagine (Asn) deamidation (1) starts with the nucleophilic attack (green arrow) of the α-amino group, in the peptide bond, to the amide group in the side chain. The cyclic succinimide intermediate is then rapidly hydrolyzed to a mixture of Aspartate (Asp) and isoAspartate (isoAsp), with the prevalent production of the latest (about 70% vs. 30%) due to the asymmetry of the succinimide [17]. The formation of isoAsp can also occur from Asp dehydration (2), with a similar nucleophilic attack, but this reaction occurs at a slower rate.
Asn deamidation is irreversible, but the isoAsp formed by the reaction can be further modified by the protein L-isoaspartyl methyltransferase (PIMT) enzyme, which converts isoAsp into Asp. Therefore, PIMT is considered a sort of repair system that limits the protein structural changes fostered by the presence of isoAsp, and indeed, due to this role, PIMT is present ubiquitously in tissues and is widely expressed in Bacteria, Archaea, and Eukarya [20].
Initially, the deamidation was thought to be purely a sort of protein damage associated to aging [18][21]. However, since both Asn and Gln residues show such instability, but are widely distributed in nature, it was proposed that deamidation can serve as a biological timer for protein turnover [21]. In fact, Asn deamidation rate depends on the amino acid sequence, so Asn deamidation could be genetically programmed.
3. Deamidation of Asn Residue in NGR Motifs: From Loss- to Gain-of-Function
Since deamidation introduces a negative charge and, in the case of isoAsp formation, changes the length of the peptide bond, this modification can induce protein loss of function. Due to local charge and conformational alterations, the functional interaction with other proteins or substrate molecules could be prevented [11][22]. For instance, in rat calmodulin the deamidation reduces the activity below 20% of the native protein [23], whereas the deamidation of the histidine-containing protein HPr impairs phosphohydrolysis and phosphotransferase activity in Bacteria [24].
Nevertheless, in some cases, Asn deamidation can result in gain of function. For example, it has been demonstrated that Asn deamidation in fibronectin is associated with increased integrin-binding properties [25]. Integrins are cell-adhesion receptors which recognize different extracellular matrix (ECM) proteins [13]. Integrins can bind multiple ligands through different specific recognition sequences [13]. One of these is the Arginine-Glycine-Aspartate (RGD) motif present in some ECM proteins like fibronectin, vitronectin, bone sialoprotein, collagen, and thrombospondin [13]. However, it has been reported that integrins can recognize another sequence within the same RGD-binding pocket: the isoDGR motif resulting from Asn deamidation of the NGR motif [26]. Indeed, structural studies showed that isoDGR binds αVβ3 integrin in reverse orientation compared to RGD, thus maintaining the tertiary structure suitable for fitting within the binding pocket [27]. It has been observed that the deamidation of Asn-263 at the NGR site of fibronectin leads to αVβ3 integrin-binding activity [25][27]. Therefore, Asn deamidation and the subsequent production of isoAsp can act as a molecular switch for integrin-binding [22].
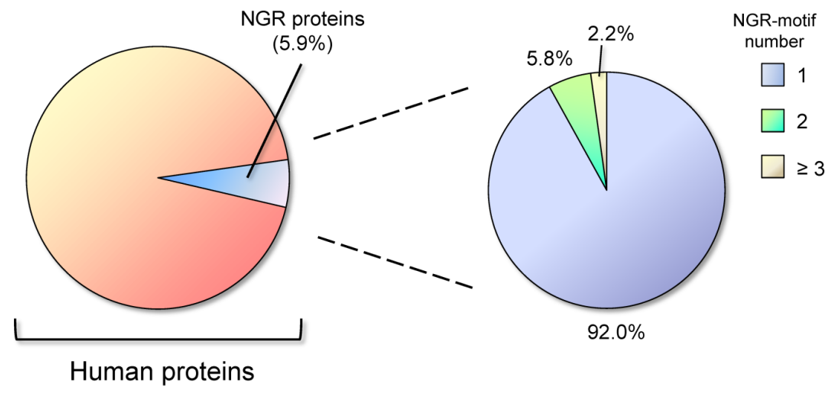
NGR motifs are also present in other proteins. A search in the Swiss-Prot databases found that about 5.02% of proteins contain the NGR-motif, but the frequency of NGR in proteins classified with the keyword “adhesion” is 17.23%. In comparison, the more investigated RGD motif is present in 6.35% of proteins [28]. We performed an up-date of this research for NGR motifs in UniProtKB/Swiss-Prot database (release 2020_05 of 07-Oct-20, 563552 entries), using Prosite tool in the Expasy bioinformatics resource portal (www.expasy.org). Excluding splice variants and restricting the analysis to the Homo sapiens database, we found that 5.9% of proteins contain the NGR motif in their sequence (1193 out of 20385). Within the total NGR-containing proteins, most of them have one NGR motif (92%), while only 8% of the proteins have two or more NGR motifs (Figure 2).
Figure 2. Distribution of Asparagine-Glycine-Arginine (NGR) motifs in human proteins. About 6% of total proteins contain the NGR motif, and the vast majority of these (92%) has one NGR motif in the sequence, while only 8% shows two or more NGR motifs.
For comparison, the integrin-binding RGD motif was found to be present in 7.7% of all human proteins, and within these, the presence of two or more RGD motifs is restricted to the 7.6%. NGR sites are therefore relatively infrequent in proteins and having two or more NGR motifs is a rare circumstance (only 0.47% of total human proteins). Performing a Gene Ontology (GO) biological process enrichment analysis by using the STRING database (www.string-db.org), we found that 14.5% of the proteins classified as to be involved in cell adhesion, contain NGR motifs; indeed, up to 18.0% of the proteins localized in the extracellular matrix hold one or more NGR site (GO Cellular Component) [29][30].
References
- Schaffert, L.N.; Carter, W.G. Do post-translational modifications influence protein aggregation in neurodegenerative diseases: A systematic review. Brain Sci. 2020, 10, doi:10.3390/brainsci10040232.
- Sambataro, F.; Pennuto, M. Post-translational Modifications and protein quality control in motor neuron and polyglutamine diseases. Mol. Neurosci. 2017, 10, 82, doi:10.3389/fnmol.2017.00082.
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease.
- Int. 2011, 58, 458–471, doi:10.1016/j.neuint.2010.12.023.
- Junqueira, S.C.; Centeno, E.G.Z.; Wilkinson, K.A.; Cimarosti, H. Post-translational modifications of Parkinson’s disease-related proteins: Phosphorylation, SUMOylation and ubiquitination. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 2001–2007, doi:10.1016/j.bbadis.2018.10.025.
- Capo, C.R.; Arciello, M.; Squitti, R.; Cassetta, E.; Rossini, P.M.; Calabrese, L.; Rossi, L. Features of ceruloplasmin in the
- cerebrospinal fluid of Alzheimer’s disease patients. Biometals 2008, 21, 367–372, doi:10.1007/s10534-007-9125-4.
- Boll, M.C.; Sotelo, J.; Otero, E.; Alcaraz-Zubeldia, M.; Rios, C. Reduced ferroxidase activity in the cerebrospinal fluid from patients with Parkinson’s disease. Lett. 1999, 265, 155–158.
- Conti, A.; Iannaccone, S.; Sferrazza, B.; De Monte, L.; Cappa, S.; Franciotta, D.; Olivieri, S.; Alessio, M. Differential expression of ceruloplasmin isoforms in the cerebrospinal fluid of amyotrophic lateral sclerosis patients. Clin. Appl. 2008, 2, 1628–1637, doi:10.1002/prca.200780081.
- Olivieri, S.; Conti, A.; Iannaccone, S.; Cannistraci, C.V.; Campanella, A.; Barbariga, M.; Codazzi, F.; Pelizzoni, I.; Magnani, G.; Pesca, M.; et al. Ceruloplasmin oxidation, a feature of Parkinson’s disease CSF, inhibits ferroxidase activity and promotes
- cellular iron retention. Neurosci. 2011, 31, 18568–18577, doi:10.1523/JNEUROSCI.3768-11.2011.
- Barbariga, M.; Curnis, F.; Spitaleri, A.; Andolfo, A.; Zucchelli, C.; Lazzaro, M.; Magnani, G.; Musco, G.; Corti, A.; Alessio, M. Oxidation-induced structural changes of ceruloplasmin foster NGR motif deamidation that promotes integrin binding and signaling. Biol. Chem. 2014, 289, 3736–3748, doi:10.1074/jbc.M113.520981.
- Barbariga, M.; Curnis, F.; Andolfo, A.; Zanardi, A.; Lazzaro, M.; Conti, A.; Magnani, G.; Volontè, M.A.; Ferrari, L.; Comi, G.; et al. Ceruloplasmin functional changes in Parkinson’s disease-cerebrospinal fluid. Neurodegener. 2015, 10, 59, doi:10.1186/s13024-015-0055-2.
- Weintraub, S.J.; Deverman, B.E. Chronoregulation by asparagine deamidation. STKE 2007, 2007, re7, doi:10.1126/stke.4092007re7.
- Barbariga, M.; Zanardi, A.; Curnis, F.; Conti, A.; Boselli, D.; Di Terlizzi, S.; Alessio, M. Ceruloplasmin oxidized and deamidated by Parkinson’s disease cerebrospinal fluid induces epithelial cells proliferation arrest and apoptosis. Rep. 2020, 10, 15507, doi:10.1038/s41598-020-72447-z.
- Plow, E.F.; Haas, T.A.; Zhang, L.; Loftus, J.; Smith, J.W. Ligand binding to integrins. Biol. Chem. 2000, 275, 21785–21788, doi:10.1074/jbc.R000003200.
- Robinson, A.B.; Rudd, C.J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Top. Cell. Regul. 1974, 8, 247–295, doi:10.1016/b978-0-12-152808-9.50013-4.
- Flatmark, T.; Sletten, K. Multiple forms of cytochrome c in the rat. Precursor-product relationship between the main
- component Cy I and the minor components Cy II and Cy 3 in vivo. Biol. Chem. 1968, 243, 1623–1629.
- Robinson, N.E.; Robinson, A.B. Prediction of primary structure deamidation rates of asparaginyl and glutaminyl peptides through steric and catalytic effects. Pept. Res. 2004, 63, 437–448, doi:10.1111/j.1399-3011.2004.00148.x.
- Geiger, T.; Clarke, S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides.
- Succinimide-linked reactions that contribute to protein degradation. Biol. Chem. 1987, 262, 785–794.
- Reissner, K.J.; Aswad, D.W. Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious
- signals? Mol. Life Sci. 2003, 60, 1281–1295, doi:10.1007/s00018-003-2287-5.
- Robinson, N.E.; Robinson, A.B. Deamidation of human proteins. Natl. Acad. Sci. USA 2001, 98, 12409–12413, doi:10.1073/pnas.221463198.
- Mishra, P.K.K.; Mahawar, M. PIMT-mediated protein repair: Mechanism and implications. Biochemistry 2019, 84, 453–463, doi:10.1134/S0006297919050018.
- Robinson, A.B.; McKerrow, J.H.; Cary, P. Controlled deamidation of peptides and proteins: An experimental hazard and a possible biological timer. Natl. Acad. Sci. USA 1970, 66, 753–757, doi:10.1073/pnas.66.3.753.
- Corti, A.; Curnis, F. Isoaspartate-dependent molecular switches for integrin-ligand recognition. Cell. Sci. 2011, 124, 515–522, doi:10.1242/jcs.077172.
- Johnson, B.A.; Langmack, E.L.; Aswad, D.W. Partial repair of deamidation-damaged calmodulin by protein carboxyl
- J. Biol. Chem. 1987, 262, 12283–12287.
- Sharma, S.; Hammen, P.K.; Anderson, J.W.; Leung, A.; Georges, F.; Hengstenberg, W.; Klevit, R.E.; Waygood, E.B. Deamidation of HPr, a phosphocarrier protein of the phosphoenolpyruvate: Sugar phosphotransferase system, involves asparagine 38 (HPr-1) and asparagine 12 (HPr-2) in isoaspartyl acid formation. Biol. Chem. 1993, 268, 17695–17704.
- Curnis, F.; Longhi, R.; Crippa, L.; Cattaneo, A.; Dondossola, E.; Bachi, A.; Corti, A. Spontaneous formation of L-isoaspartate and gain of function in fibronectin. Biol. Chem. 2006, 281, 36466–36476, doi:10.1074/jbc.M604812200.
- Takahashi, S.; Leiss, M.; Moser, M.; Ohashi, T.; Kitao, T.; Heckmann, D.; Pfeifer, A.; Kessler, H.; Takagi, J.; Erickson, H.P.; et al. The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. Cell. Biol. 2007, 178, 167–178, doi:10.1083/jcb.200703021.
- Spitaleri, A.; Mari, S.; Curnis, F.; Traversari, C.; Longhi, R.; Bordignon, C.; Corti, A.; Rizzardi, G.P.; Musco, G. Structural basis for the interaction of isoDGR with the RGD-binding site of alphavbeta3 integrin. Biol. Chem. 2008, 283, 19757–19768, doi:10.1074/jbc.M710273200.
- Corti, A.; Curnis, F.; Arap, W.; Pasqualini, R. The neovasculature homing motif NGR: More than meets the eye. Blood 2008, 112, 2628–2635, doi:10.1182/blood-2008-04-150862.
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338, doi:10.1093/nar/gky1055.
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in
- genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613, doi:10.1093/nar/gky1131.