| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Simon Cocklin | + 2227 word(s) | 2227 | 2021-02-07 04:05:36 | | | |
| 2 | Bruce Ren | Meta information modification | 2227 | 2021-02-20 07:12:04 | | |
Video Upload Options
The capsid (CA) protein of the human immunodeficiency virus type 1 (HIV-1) is an essential structural component of a virion and facilitates many crucial life cycle steps through interactions with host cell factors. Capsid shields the reverse transcription complex from restriction factors while it enables trafficking to the nucleus by hijacking various adaptor proteins, such as FEZ1 and BICD2. In addition, the capsid facilitates the import and localization of the viral complex in the nucleus through interaction with NUP153, NUP358, TNPO3, and CPSF-6. In the later stages of the HIV-1 life cycle, CA plays an essential role in the maturation step as a constituent of the Gag polyprotein. In the final phase of maturation, Gag is cleaved, and CA is released, allowing for the assembly of CA into a fullerene cone, known as the capsid core. The fullerene cone consists of ~250 CA hexamers and 12 CA pentamers and encloses the viral genome and other essential viral proteins for the next round of infection. As research continues to elucidate the role of CA in the HIV-1 life cycle and the importance of the capsid protein becomes more apparent, CA displays potential as a therapeutic target for the development of HIV-1 inhibitors.
1. Introduction
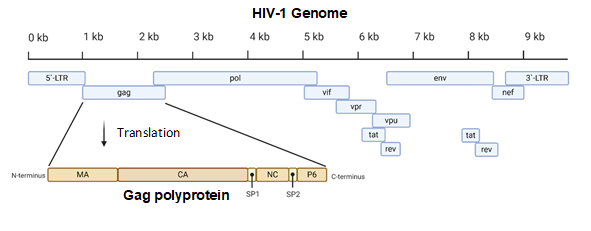
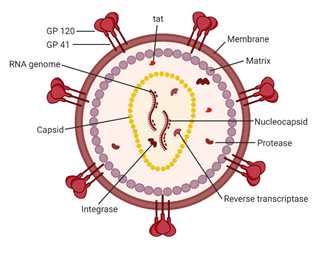
Acquired immunodeficiency syndrome (AIDS) affected approximately 38 million people in 2019. The etiologic agent for AIDS is the human immunodeficiency virus (HIV) [1]. While HIV is categorized into two subgroups, type 1 and type 2, HIV type 1 (HIV-1) is the most prevalent cause of AIDS worldwide [2]. It is an enveloped virus containing a 9.8kb positive-sense RNA genome (Figure 1) that codes for three polyproteins (Gag, Pol, and Env) and six accessory proteins (Tat, Rev, Nef, Vpr, Vif, and Vpu) [3]. HIV-1 targets human cells presenting the CD4 receptor and CCR5 or CXCR4 co-receptors, such as T helper cells and microglial cells [4][5]. It penetrates the host cell through receptor-mediated entry, which results in the viral core entering the cytoplasm of the host cell [6]. The capsid core is a fullerene-like cone made of the capsid (CA) portion of the Gag polyprotein. The core contains the viral genome and viral proteins essential for replication, such as integrase and reverse transcriptase (Figure 2) [7]. The CA protein is essential in both the early and late stages of the HIV-1 life cycle, with many host cell factors currently identified as direct binding partners [7].
Figure 1. A diagram of the 9.8 kb HIV-1 genome. The Gag portion of the genome is transcribed into the Gag polyprotein, consisting of the matrix (MA), capsid (CA), nucleocapsid (NC), P6, and two spacer peptides (SP1 and SP2). During maturation, the polyprotein is cleaved into its constituent parts. Image created with BioRender.com.
Figure 2. A schematic of the HIV-1 virion. Envelope proteins, GP41 and GP120, surround the host-derived membrane surface, which is lined internally with a layer of matrix protein. Inside the virion are viral proteins and the CA core containing the HIV-1 genome and proteins essential for infection. Image created with BioRender.com.
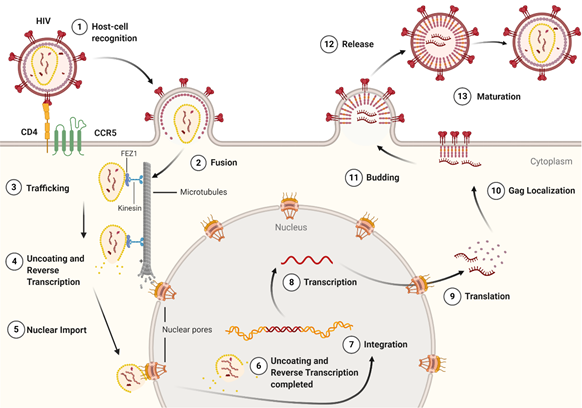
The life cycle of HIV-1 (Figure 3) can be broken down into two stages: early and late. The early-stage begins with an infectious virion binding to the host cell and ends with the integration of the viral genome into the host DNA. The late stage of the life cycle is the period from post-integration until viral maturation [8]. The HIV-1 life cycle's early stage begins with the virion’s glycoprotein complex, Env, interacting with the CD4 receptor and the CCR5 or CXCR4 co-receptors on the host cell [9]. This recognition event initiates a cascade of conformational rearrangements that results in viral fusion, where the viral core is released into the cytoplasm of the host cell [9]. The complex of the capsid protein and its contents is referred to as the reverse-transcription complex (RTC). From here, reverse transcription must occur within the core. During reverse transcription, capsid begins uncoating and trafficking to the nucleus for import and integration [10]. Once the RTC has entered the nucleus, it is referred to as the pre-integration complex (PIC) The processes of uncoating and reverse transcription are then completed after nuclear import and before nuclear integration [11][12][13]. The capsid is responsible for localizing the PIC to transcriptionally active sites on chromatin and facilitating the integration of the viral transcripts into the host genome [14][15][16]. The viral genes are then transcribed by the host cell and exported from the nucleus. Export is followed by localization of the Gag protein to the plasma membrane through a myristoyl group at the amino-terminus of Gag [17]. Localization is followed by an immature virion budding, with intact Gag polyproteins coating the host-derived viral membrane [18]. The final step of the viral life cycle is maturation, which results in a fully infectious virion. The maturation step is facilitated by the viral protease that cleaves the polyprotein into its smaller, functional constituents [19]. In this step, the viral capsid is formed and assembles into the fullerene-cone, forming a mature and fully infectious HIV-1 virion. At this stage, the newly formed and matured virion can restart the HIV-1 life cycle, infecting another host cell [19].
Figure 3. The life cycle of the HIV-1 virus. The early-stage begins with the recognition of host cell receptors (1), resulting in the fusion of the virus and release of the viral core into the cytoplasm of the host cell (2). This is followed by the trafficking of the core through the cytoplasm (3) as reverse transcription and uncoating begins to take place (4). Once at the nuclear pore, the viral contents are imported into the nucleus and localized (5) to transcriptionally active chromatin while uncoating and reverse transcription are completed (6). Following uncoating and reverse transcription, integration occurs (7). After the viral genome is integrated into the host cell, viral genes are transcribed (8) and translated (9) into the Gag polyprotein. The Gag polyprotein then localizes to the host cell membrane (10), where budding occurs (11), followed by the release of an immature virion (12). The final step in the HIV-1 lifecycle is maturation (13), where the viral protease cleaves the Gag polyprotein into its constituent, functional proteins. Image created with BioRender.com.
Continued studies into the life cycle of HIV-1 have revealed that CA is essential in infection and replication. It provides structure and protection to the RTC and PIC while allowing dNTP diffusion for reverse transcription [20][21]. Additionally, it facilitates retrograde movement in the cytoplasm, nuclear import, and localization [22][23][24][25][26][27]. Since CA is essential in the HIV-1 lifecycle, it is a promising target for future research into HIV-1 inhibitors [28][29][30][31][32][33][34].
2. Structure of the HIV-1 Capsid
2.1. Gag Polyprotein
The proteins of HIV-1 are transcribed and translated as polyproteins, which are long peptides that are cleaved into smaller, functional units by the viral protease [35]. The CA protein of HIV-1 is translated as a constituent of the larger Gag polyprotein (Figure 1) that contains many integral viral components [36]. This intact Gag protein is essential in the late stages of the viral life cycle. It is responsible for the localization of viral components to the host cell plasma membrane through a myristoyl group located at its amino-terminus. Following budding, the virus is still immature because the Gag polyprotein remains whole. It is not until cleavage by the viral protease that the polyprotein is separated into its individual constituents, and a mature, infectious virion is formed [37].
2.2. CA Monomer: The Building Block of the Capsid Core
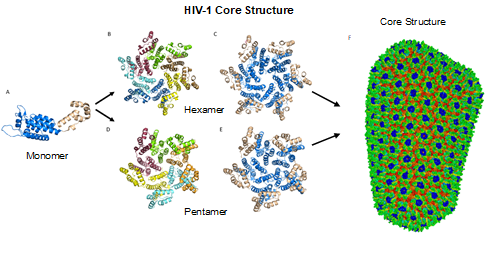
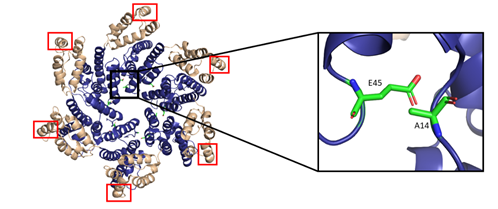
The most basic component of the capsid core is the CA monomer (Figure 4A), a highly conserved and genetically fragile protein [38]. The CA monomer is a 231-residue long protein with two major alpha-helical domains, the N-terminal and C-terminal domains (CA-NTD and CA-CTD, respectively) that are connected by a five-residue linker (residues 146–150) [39]. While a virion is in its immature form, CA is a constituent of the Gag polyprotein, linked to the matrix (MA) domain at its N-terminus and the spacer peptide 1 (SP1) domain of the polyprotein at its C-terminus (Figure 1). After budding and during maturation, the Gag polyprotein undergoes proteolytic cleavage, separating it from the MA and SP1 domains. Interactions then occur between CA residues Ala14 and Glu45 of adjacent monomers, allowing the monomers to form larger oligomers (Figure 7) [40]. One study that examined CA dynamics determined that the protein exists in an equilibrium of monomers and dimers. In this equilibrium, the orientation of the NTD and CTD exists in a range of different states. This equilibrium was suggested to account for the pentamer/hexamer configuration seen in the fully assembled CA core, with the variability of the NTD of the dimer determining the pentamer/hexamer pattern of assembly in the finished core [41].
Figure 4. HIV-1 CA monomers oligomerize into hexamers and pentamers that assemble to form the capsid fullerene-cone core. (A) Shows the CA monomer with the amino-terminus colored blue and the carboxyl-terminus colored tan (PDB 3H47). (B) Shows the CA hexamer with each subunit colored differently (PDB 3H47). (C) Shows the hexamer with the amino-terminus of each subunit colored blue and the carboxyl terminus colored tan (PDB 3H47). (D) Shows the CA pentamer with each subunit colored differently (PDB 3P05). (E) Shows the pentamer with the amino-terminus of each subunit colored blue and the carboxyl terminus colored tan (PDB 3P05). (F) The complete capsid core structure containing approximately 250 hexamer oligomers and 12 pentamer oligomers (PDB 3J3Q). Image created with the PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC.
2.3. CA Oligomers: Pentamers and Hexamers
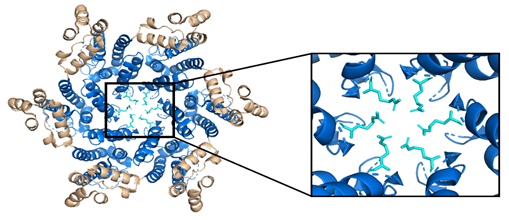
While the HIV-1 CA monomers may form various curved oligomers, the most abundant oligomer in the mature cone is the hexamer (Figure 4B, 4C). Approximately 250 hexamers and precisely 12 pentamers (Figure 4D,4E) form the fullerene-like cone lattice of the viral capsid core (Figure 4F). The hexamers are structured with the N-terminal domain forming a central, stable core and the C-terminal domains forming a flexible outer ring. The center of the hexamer core, referred to as the R18 pore, is lined with six arginine residues and a “molecular-iris” formed from the N-terminal β-hairpin (Figure 5). This pore is a size-selective, positively charged channel that allows for the diffusion of nucleotides into the capsid core while preventing access to nucleases, host cell restriction factors, and sensors.
Figure 5. The HIV-1 CA hexamer with the R18 ring expanded. The amino-terminus is colored blue, the carboxyl-terminus is colored tan, and each subunit's R18 residue is colored in cyan. This is referred to as the R18 ring. This pore allows for the diffusion of materials needed for reverse transcription while sequestering reverse transcription machinery from host cell restriction factors. (PDB 3H47) Image created with the PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC.
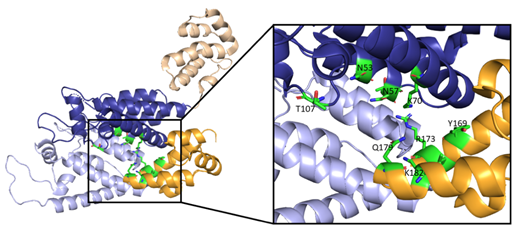
Along with the R18 pore, there is a crucial interprotomer pocket (Figure 6) that forms between hexameric subunits and is generated by the NTD of one subunit interacting with the CTD of another subunit. This NTD-CTD interprotomer interface is present in the mature HIV-1 capsid and is critical for proper capsid assembly, stability, and host cell factor recognition. There is evidence that this pocket binds host cell factors involved in nuclear import, suggesting that intact CA oligomers are imported into the nucleus during nuclear entry of the pre-integration complex (PIC) [42]. Due to these roles, the interprotomer interface has been used as a target for small-molecule inhibition [43].
Additional interactions involved in the polymerization of capsid monomers include the NTD-NTD interactions that promote the formation of hexamers and CTD-CTD interfaces that connects hexameric units into the conical lattice [44]. The NTD-NTD interface involves helices 1 and 3 of one monomer and helix 2 of the adjacent monomer [40]. Conversely, the CTD-CTD interface links two neighboring capsid subunits belonging to different hexamers and is defined by helix 9 of the adjacent monomers [44].
Figure 6. Here, two adjacent CA protomers of the CA hexamer form a functional interprotomer pocket. N-terminal domains (NTDs) are colored in shades of blue, and C-terminal domains (CTDs) are colored in shades of orange. Interacting domains are marked in dark blue and bright orange. Interacting residues within the interaction interface are magnified and colored in green. (PDB 6MQA) Image created with the PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC.
2.4. The Fullerene Cone
The hexamers and pentamers associate into the curved lattice that forms the mature capsid core. Electron microscopy has demonstrated that the natural viral core exhibits cone angles of ~19°, indicating that the capsid core of HIV-1 is structured as a fullerene cone, with the rigid rotation between the pentamers and hexamers appearing to generate the continuous curve of the fullerene cone [45][46]. However, both perfect and aberrant fullerene cones have been observed after maturation and are believed to be caused by the de novo capsid assembly that occurs after proteolytic cleavage of the Gag polyprotein [47]. The fullerene cone is an important structure, as it protects the viral genome from a wide range of host cell factors that would otherwise restrict infection. Mutagenesis studies revealed that residues within the interprotomer pocket and pentamer/hexamer interface are essential, with disruption of those interactions leading to decreased capsid assembly, stability, and viral infectivity [48][49][50][51][52][53]. Specifically, the hydrophobic residues in the three-helix bundle center within the CA-CTD are crucial for hexamer/pentamer association (Figure 7). This again emphasizes the importance of a stable capsid core since instability in the core has been shown to cause a loss of the viral genome and integration [54].
Figure 7. The HIV-1 CA hexamer with regions important for higher-order oligomeric assembly are highlighted. The amino-terminus is colored blue, and the carboxyl-terminus is colored tan. Residues A14 and E45 between monomeric subunits are magnified and colored green. The regions of hydrophobic residues within the three-helix bundle center are marked with red boxes. (PDB 6MQA) Image created with the PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC.
References
- Clements, J.E.; Zink, M.C. Molecular biology and pathogenesis of animal lentivirus infections. Clin. Microbiol. Rev. 1996, 9, 100–117, doi:10.1128/CMR.9.1.100-117.1996.
- Bbosa, N.; Kaleebu, P.; Ssemwanga, D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS 2019, 14, 153–160, doi:10.1097/COH.0000000000000534.
- German Advisory Committee Blood, Subgroup ‘Assessment of Pathogens Transmissible by Blood'. Human Immunodeficiency Virus (HIV). Transfus Med. Hemother. 2016, 43, 203–222, doi:10.1159/000445852.
- Cenker, J.J.; Stultz, R.D.; McDonald, D. Brain Microglial Cells Are Highly Susceptible to HIV-1 Infection and Spread. AIDS Res. Hum. Retrovir. 2017, 33, 1155–1165, doi:10.1089/AID.2017.0004.
- Douek, D.C.; Brenchley, J.M.; Betts, M.R.; Ambrozak, D.R.; Hill, B.J.; Okamoto, Y.; Casazza, J.P.; Kuruppu, J.; Kunstman, K.; Wolinsky, S.; et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature 2002, 417, 95–98, doi:10.1038/417095a.
- Hunter, E. Viral Entry and Receptors; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997.
- Novikova, M.; Zhang, Y.; Freed, E.O.; Peng, K. Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol. Sin. 2019, 34, 119–134, doi:10.1007/s12250-019-00095-3.
- Campbell, E.M.; Hope, T.J. Live cell imaging of the HIV-1 life cycle. Trends Microbiol. 2008, 16, 580–587, doi:10.1016/j.tim.2008.09.006.
- Moore, J.P.; Trkola, A.; Dragic, T. Co-receptors for HIV-1 entry. Curr. Opin. Immunol. 1997, 9, 551–562, doi:10.1016/s0952-7915(97)80110-0.
- Cosnefroy, O.; Murray, P.J.; Bishop, K.N. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 2016, 13, 58, doi:10.1186/s12977-016-0292-7.
- Selyutina, A.; Persaud, M.; Lee, K.; KewalRamani, V.; Diaz-Griffero, F. Nuclear Import of the HIV-1 Core Precedes Reverse Transcription and Uncoating. Cell Rep. 2020, 32, 108201, doi:10.1016/j.celrep.2020.108201.
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493, doi:10.1073/pnas.1920631117.
- Dharan, A.; Bachmann, N.; Talley, S.; Zwikelmaier, V.; Campbell, E.M. Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 2020, 5, 1088–1095, doi:10.1038/s41564-020-0735-8.
- Dharan, A.; Talley, S.; Tripathi, A.; Mamede, J.I.; Majetschak, M.; Hope, T.J.; Campbell, E.M. KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection. PLoS Pathog. 2016, 12, e1005700, doi:10.1371/journal.ppat.1005700.
- Krishnan, L.; Matreyek, K.A.; Oztop, I.; Lee, K.; Tipper, C.H.; Li, X.; Dar, M.J.; Kewalramani, V.N.; Engelman, A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 2010, 84, 397–406, doi:10.1128/JVI.01899-09.
- Larue, R.; Gupta, K.; Wuensch, C.; Shkriabai, N.; Kessl, J.J.; Danhart, E.; Feng, L.; Taltynov, O.; Christ, F.; Van Duyne, G.D.; et al. Interaction of the HIV-1 intasome with transportin 3 protein (TNPO3 or TRN-SR2). J. Biol. Chem. 2012, 287, 34044–34058, doi:10.1074/jbc.M112.384669.
- Bryant, M.; Ratner, L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 1990, 87, 523–527, doi:10.1073/pnas.87.2.523.
- Usami, Y.; Popov, S.; Popova, E.; Inoue, M.; Weissenhorn, W.; H., G.G. The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 2009, 37, 181–184, doi:10.1042/BST0370181.
- De Marco, A.; Muller, B.; Glass, B.; Riches, J.D.; Krausslich, H.G.; Briggs, J.A. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010, 6, e1001215, doi:10.1371/journal.ppat.1001215.
- Jacques, D.A.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 2016, 536, 349–353, doi:10.1038/nature19098.
- Song, G. Uncovering the release mechanism of nucleotide import by HIV-1 capsid. Phys. Biol. 2020, 18, 016004, doi:10.1088/1478-3975/abbf32.
- Achuthan, V.; Perreira, J.M.; Ahn, J.J.; Brass, A.L.; Engelman, A.N. Capsid-CPSF6 interaction: Master regulator of nuclear HIV-1 positioning and integration. J. Life Sci. 2019, 1, 39–45, doi:10.36069/jols/20190604.
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404 e398, doi:10.1016/j.chom.2018.08.002.
- Buffone, C.; Martinez-Lopez, A.; Fricke, T.; Opp, S.; Severgnini, M.; Cifola, I.; Petiti, L.; Frabetti, S.; Skorupka, K.; Zadrozny, K.K.; et al. Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells. J. Virol. 2018, 92, doi:10.1128/JVI.00648-18.
- Bukrinsky, M.I.; Haffar, O.K. HIV-1 nuclear import: In search of a leader. Front. Biosci. 1999, 4, D772–D781, doi:10.2741/bukrinsky.
- Carnes, S.K.; Zhou, J.; Aiken, C. HIV-1 Engages a Dynein-Dynactin-BICD2 Complex for Infection and Transport to the Nucleus. J. Virol. 2018, 92, doi:10.1128/JVI.00358-18.
- Dharan, A.; Opp, S.; Abdel-Rahim, O.; Keceli, S.K.; Imam, S.; Diaz-Griffero, F.; Campbell, E.M. Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc. Natl. Acad. Sci. USA 2017, 114, E10707–E10716, doi:10.1073/pnas.1712033114.
- Abdurahman, S.; Hoglund, S.; Hoglund, A.; Vahlne, A. Mutation in the loop C-terminal to the cyclophilin A binding site of HIV-1 capsid protein disrupts proper virus assembly and infectivity. Retrovirology 2007, 4, 19, doi:10.1186/1742-4690-4-19.
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364, doi:10.1126/science.abb4808.
- Carnes, S.K.; Sheehan, J.H.; Aiken, C. Inhibitors of the HIV-1 capsid, a target of opportunity. Curr .Opin. HIV AIDS 2018, 13, 359–365, doi:10.1097/COH.0000000000000472.
- Singh, K.; Gallazzi, F.; Hill, K.J.; Burke, D.H.; Lange, M.J.; Quinn, T.P.; Neogi, U.; Sonnerborg, A. GS-CA Compounds: First-In-Class HIV-1 Capsid Inhibitors Covering Multiple Grounds. Front. Microbiol. 2019, 10, 1227, doi:10.3389/fmicb.2019.01227.
- Sun, L.; Dick, A.; Meuser, M.E.; Huang, T.; Zalloum, W.A.; Chen, C.H.; Cherukupalli, S.; Xu, S.; Ding, X.; Gao, P.; et al. Design, Synthesis, and Mechanism Study of Benzenesulfonamide-Containing Phenylalanine Derivatives as Novel HIV-1 Capsid Inhibitors with Improved Antiviral Activities. J. Med. Chem. 2020, 63, 4790–4810, doi:10.1021/acs.jmedchem.0c00015.
- Thenin-Houssier, S.; de Vera, I.M.; Pedro-Rosa, L.; Brady, A.; Richard, A.; Konnick, B.; Opp, S.; Buffone, C.; Fuhrmann, J.; Kota, S.; et al. Ebselen, a Small-Molecule Capsid Inhibitor of HIV-1 Replication. Antimicrob. Agents Chemother. 2016, 60, 2195–2208, doi:10.1128/AAC.02574-15.
- Xu, J.P.; Francis, A.C.; Meuser, M.E.; Mankowski, M.; Ptak, R.G.; Rashad, A.A.; Melikyan, G.B.; Cocklin, S. Exploring Modifications of an HIV-1 Capsid Inhibitor: Design, Synthesis, and Mechanism of Action. J. Drug Des. Res. 2018, 5, 1070.
- Erickson-Viitanen, S.; Manfredi, J.; Viitanen, P.; Tribe, D.E.; Tritch, R.; Hutchison, C.A., 3rd; Loeb, D.D.; Swanstrom, R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: Sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 1989, 5, 577–591, doi:10.1089/aid.1989.5.577.
- Su, C.T.; Kwoh, C.K.; Verma, C.S.; Gan, S.K. Modeling the full length HIV-1 Gag polyprotein reveals the role of its p6 subunit in viral maturation and the effect of non-cleavage site mutations in protease drug resistance. J. Biomol. Struct. Dyn. 2018, 36, 4366–4377, doi:10.1080/07391102.2017.1417160.
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646, doi:10.1038/nature12162.
- Rihn, S.J.; Wilson, S.J.; Loman, N.J.; Alim, M.; Bakker, S.E.; Bhella, D.; Gifford, R.J.; Rixon, F.J.; Bieniasz, P.D. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013, 9, e1003461, doi:10.1371/journal.ppat.1003461.
- Jiang, J.; Ablan, S.D.; Derebail, S.; Hercik, K.; Soheilian, F.; Thomas, J.A.; Tang, S.; Hewlett, I.; Nagashima, K.; Gorelick, R.J.; et al. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology 2011, 421, 253–265, doi:10.1016/j.virol.2011.09.012.
- Pornillos, O.; Ganser-Pornillos, B.K.; Kelly, B.N.; Hua, Y.; Whitby, F.G.; Stout, C.D.; Sundquist, W.I.; Hill, C.P.; Yeager, M. X-ray structures of the hexameric building block of the HIV capsid. Cell 2009, 137, 1282–1292, doi:10.1016/j.cell.2009.04.063.
- Deshmukh, L.; Schwieters, C.D.; Grishaev, A.; Ghirlando, R.; Baber, J.L.; Clore, G.M. Structure and dynamics of full-length HIV-1 capsid protein in solution. J. Am. Chem. Soc. 2013, 135, 16133–16147, doi:10.1021/ja406246z.
- Bhattacharya, A.; Alam, S.L.; Fricke, T.; Zadrozny, K.; Sedzicki, J.; Taylor, A.B.; Demeler, B.; Pornillos, O.; Ganser-Pornillos, B.K.; Diaz-Griffero, F.; et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625–18630, doi:10.1073/pnas.1419945112.
- Yufenyuy, E.L.; Aiken, C. The NTD-CTD intersubunit interface plays a critical role in assembly and stabilization of the HIV-1 capsid. Retrovirology 2013, 10, 29, doi:10.1186/1742-4690-10-29.
- Bocanegra, R.; Alfonso, C.; Rodríguez-Huete, A.; Fuertes, Miguel, Á.; Jiménez, M.; Rivas, G.; Mateu, Mauricio, G. Association Equilibrium of the HIV-1 Capsid Protein in a Crowded Medium Reveals that Hexamerization during Capsid Assembly Requires a Functional C-Domain Dimerization Interface. Biophys. J. 2013, 104, 884–893, doi:https://doi.org/10.1016/j.bpj.2012.12.035.
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and analysis of conical models for the HIV-1 core. Science 1999, 283, 80–83, doi:10.1126/science.283.5398.80.
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nature 2011, 469, 424–427, doi:10.1038/nature09640.
- Mattei, S.; Glass, B.; Hagen, W.J.; Krausslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437, doi:10.1126/science.aah4972.
- von Schwedler, U.K.; Stray, K.M.; Garrus, J.E.; Sundquist, W.I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003, 77, 5439–5450, doi:10.1128/jvi.77.9.5439-5450.2003.
- Hulme, A.E.; Kelley, Z.; Okocha, E.A.; Hope, T.J. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J. Virol. 2015, 89, 643–651, doi:10.1128/jvi.03043-14.
- Tang, S.; Murakami, T.; Agresta, B.E.; Campbell, S.; Freed, E.O.; Levin, J.G. Human Immunodeficiency Virus Type 1 N-Terminal Capsid Mutants That Exhibit Aberrant Core Morphology and Are Blocked in Initiation of Reverse Transcription in Infected Cells. J. Virol. 2001, 75, 9357, doi:10.1128/JVI.75.19.9357-9366.2001.
- Tang, S.; Murakami, T.; Cheng, N.; Steven, A.C.; Freed, E.O.; Levin, J.G. Human immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 2003, 77, 12592–12602, doi:10.1128/jvi.77.23.12592-12602.2003.
- Fitzon, T.; Leschonsky, B.; Bieler, K.; Paulus, C.; Schröder, J.; Wolf, H.; Wagner, R. Proline residues in the HIV-1 NH2-terminal capsid domain: Structure determinants for proper core assembly and subsequent steps of early replication. Virology 2000, 268, 294–307, doi:10.1006/viro.1999.0178.
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a Human Immunodeficiency Virus Type 1 Core of Optimal Stability Is Crucial for Viral Replication. J. Virol. 2002, 76, 5667, doi:10.1128/JVI.76.11.5667-5677.2002.
- Eschbach, J.E.; Elliott, J.L.; Li, W.; Zadrozny, K.K.; Davis, K.; Mohammed, S.J.; Lawson, D.Q.; Pornillos, O.; Engelman, A.N.; Kutluay, S.B. Capsid lattice destabilization leads to premature loss of the viral genome and integrase enzyme during HIV-1 infection. J. Virol. 2020, 10.1128/JVI.00984-20, doi:10.1128/JVI.00984-20.