
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Anna Kurzyńska-Kokorniak | + 2566 word(s) | 2566 | 2021-02-04 04:20:16 | | | |
| 2 | Peter Tang | Meta information modification | 2566 | 2021-02-22 13:28:59 | | |
Video Upload Options
Ribonuclease Dicer belongs to the family of RNase III endoribonucleases, the enzymes that specifically hydrolyze phosphodiester bonds found in double-stranded regions of RNAs. Dicer enzymes are mostly known for their essential role in the biogenesis of small regulatory RNAs. A typical Dicer-type RNase consists of a helicase domain, a domain of unknown function (DUF283), a PAZ (Piwi-Argonaute-Zwille) domain, two RNase III domains, and a double-stranded RNA binding domain; however, the domain composition of Dicers varies among species. Dicer and its homologues developed only in eukaryotes; nevertheless, the two enzymatic domains of Dicer, helicase and RNase III, display high sequence similarity to their prokaryotic orthologs. Evolutionary studies indicate that a combination of the helicase and RNase III domains in a single protein is a eukaryotic signature and is supposed to be one of the critical events that triggered the consolidation of the eukaryotic RNA interference.
1. Introduction
The gene encoding for Dicer ribonuclease, as reported by Hansen et al. [1], was first revealed in 1994 by Bass et al. in a screen of the Xenopus ovary cDNA expression library for double-stranded RNA (dsRNA) binding proteins (dsRBPs) [2]. Later, in 1999, Provost et al. published the first literature record related to human Dicer (hDicer) [3]. The authors isolated hDicer cDNA clones from a yeast two-hybrid screen, using as a bait, 5-lipoxygenase, a pivotal protein in cellular leukotriene synthesis. Analysis of the predicted amino acid sequence of the newly identified protein revealed its high homology to a hypothetical helicase K12H4.8 from Caenorhabditis elegans as well as the presence of a ribonuclease III (RNase III) signature motif and a dsRNA binding domain (dsRBD) [3]. A year later, in 2000, a detailed description of the DICER1 gene was published [4], at that time under the name “HERNA” (helicase with RNase motif), because the protein product of the gene contained conservative evolutionary motifs characteristic of the ATP-dependent RNA helicase, i.e., ATP binding motif and DEXD/H-box (Asp-Glu-X-Asp/His), and RNA binding motifs [4]. Expression of HERNA was shown to be ubiquitous and tissue-specific [4]. The name “Dicer” was first introduced into the scientific world in 2001 by Bernstein, Caudy, Hammond and Hannon [5], and it derived from the ability of the enzyme to “dice”, i.e., digest dsRNA into uniformly sized small RNAs, ~22 nucleotides (nt) in length. As Dicer displayed specificity for dsRNAs, it was assigned to the family of RNase III endoribonucleases [5].
Relating to the plant kingdom, the first literature record of Dicer goes back to 1999, when Jacobsen et al. identified in Arabidopsis thaliana the CARPEL FACTORY (CAF) gene, which played a role in floral meristem determinacy [6]. The CAF gene encoded a putative protein of 1909 amino acids containing an amino (N)-terminal DExH/DEAD-box type RNA helicase domain and a carboxy (C)-terminal RNaseIII-like domain followed by a dsRNA binding domain [6]. In those days, as mentioned above, a highly similar protein of an unknown function was found to be encoded by a fungal and an animal genome, indicating that the CAF-type proteins could play a role in many eukaryotic organisms. Moreover, the structure of CAF protein suggested that it could act as an RNA processing enzyme. Later, in 2002, Reinhart et al. made the observation that the plant microRNA (miRNA) accumulation is dependent on the CAF protein [7]. Nowadays, the CAF gene is referred to as DICER-LIKE1 (DCL1), which encodes well known endonuclease that produces plant miRNAs [8]. Identification of three other genes similar to CAF in A. thaliana led to the recognition of additional DICER family members: DCL2, DCL3 and DCL4 [9].
RNase III specifically hydrolyzes phosphodiester bonds found in double-stranded regions of RNAs and generate products having 5′-phosphate, 3′-hydroxyl, and 2-nt 3′-overhangs [10]. Based on the domain organization and biological functions, the RNase III family can be divided into three subfamilies (Figure 1). The first subfamily includes bacterial RNase III and yeast Rnt1, which contain only one RNase III domain and one dsRBD domain. RNase IIIs of this subfamily form homodimers to assemble a dsRNA-cleavage center [11]. The bacterial RNase III and yeast Rnt1 are essential for processing of ribosomal RNA precursors [12]. Moreover, Rnt1 functions in the maturation of both small nuclear RNAs [13][14] and small nucleolar RNAs [15], in processing of mRNA [16], and mRNA quality control [17]. The second subfamily of RNase III is represented by enzymes containing two RNase III domains and a single dsRBD in the C-terminal region, e.g., ribonuclease Drosha. Drosha is the core nuclease initiating processing of a primary miRNA precursor (pri-miRNA) in the nucleus. Products of Drosha cleavage are 50–70-nt pre-miRNAs with hairpin structures [18]. Drosha is also involved in mRNA processing [19] and, presumably, in ribosomal RNA maturation [20][21]. Finally, the third subfamily of RNase III comprises Dicer-type enzymes. Dicer-type RNases consist of two RNase III domains and a dsRBD in the C-terminal region, and they also include helicase domain, a DUF283 (Domain of Unknown Function) and a PAZ (Piwi–Argonaute–Zwille) domain in the N-terminal region. Nevertheless, there are a few examples of Dicers that lack some of these domains, e.g., the Dicer from Giardia intestinalis, which does not possess helicase, DUF283, and dsRBD domains [10], A. thaliana DICER-LIKE 3 (DCL3) protein, which lacks a DUF283 domain [22], or the green alga Chlamydomonas reinhardtii DCL3, which lacks the PAZ domain [23]. The second and third subfamily members use intramolecular dimerization of their two RNase III domains to form a cleavage center. Dicer ribonucleases are mostly known for their essential role in the biogenesis of small regulatory RNAs, such as miRNAs or small interfering RNAs (siRNAs). They recognize and cleave pre-miRNAs or dsRNAs into functional 21–23-nt miRNAs or siRNAs, respectively.
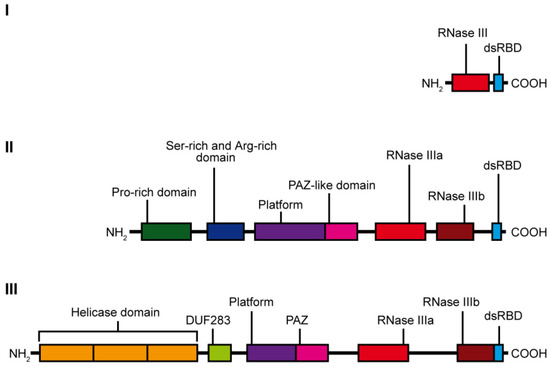
Figure 1. Scheme showing the domain organization of the individual classes of the RNase III family enzymes: class I—represented by E. coli ribonuclease III, class II—represented by human Drosha ribonuclease and class III—represented by hDicer ribonuclease.
2. Evolution of Dicer-Type Proteins
Evolutionary studies indicate that Dicer and its homologues developed only in eukaryotes, and the Dicer gene underwent duplication during the emergence of the first multicellular organisms [24]. Then, Dicer genes differentiated independently in the animal, plant and fungal kingdoms [24]. Animals developed two genes coding for Dicer proteins, Dicer-1 and Dicer-2. Dicer-1 is responsible for pre-miRNA recognition and miRNA production, while Dicer-2 cleaves dsRNA to generate siRNA [25]. One of the best characterized Dicer-2 proteins in the animal kingdom is the insect Dicer-2 which, by cleaving viral dsRNAs, plays an important role in the response to viral infections [26][27]. Dicer-2 was subsequently lost from lineages that developed complex immune systems, such as vertebrates [24]. Moreover, the Dicer gene was lost from some protozoan parasites and some fungi, e.g., the model organism Saccharomyces cerevisiae and other closely related yeasts [28][29]. In the plant kingdom, at least four genes encoding Dicer-type enzymes developed [24]. Plant Dicers are termed DICER-LIKE (DCL) proteins. DCL1 is involved in the production of miRNAs, typically 21-nt long, from primary transcripts that contain stem-loop structures, while DCL2, DCL3 and DCL4 are involved in siRNA production [30]. Precisely, DCL2 produces 22-nt siRNAs which can silence transgenes in the plant response to viral infections, and similarly to its insect homolog Dicer-2, is a part of the antiviral defense system [31][32]. DCL3 generates 24-nt siRNA, which participates in chromatin modifications, thus influencing the chromatin structure [33]. DCL4 is responsible for the formation of trans-acting siRNAs (tasiRNAs) [34]. DCL4 is also involved in the antiviral response [31]. Some plants encode more than four DCL proteins; for example, in Medicago truncatula, six DICER-LIKE protein genes are expressed: DCL1, DCL2, DCL3, DCL4 and two DCL2 homologues, including the variant that encodes a truncated version of the DCL2 protein [35]. Plant DCL proteins form a protein family, whose diversification time dates to the emergence of mosses (Physcomitrella patens) [36]. DCL proteins are ubiquitously but not evenly expressed in plant tissues, depending on developmental stages and response to stresses [36].
It is also important to mention DICER proteins encoded by algae. Chlamydomonas reinhardtii is a single-cell green alga, which has a complex RNA silencing machinery involving three DCL proteins (DCL1-3) and three Argonaute proteins (AGO1-3) [37][38]. The miRNAs silencing system in algae is distinct from that of land plants and even may have common features with the miRNA silencing mechanism found in animals. DCL3 of C. reinhardtii is known to process the majority of miRNA, although it lacks the PAZ domain [23]. Another unique feature of DCL3 is a proline-rich domain on the N-terminal side of the RNase III motifs, though a similar domain is also present in a related protein, a ribonuclease Drosha that also lacks the PAZ domain [23][39]. Considering these features, it could be possible that C. reinhardtii DCL3 performs the function of Dicer and Drosha, being involved in several stages of miRNA biogenesis [23]. Plant and animal miRNA pathways are very different, thus it is likely that they evolved separately from an ancestral RNA silencing pathway with Dicer proteins [40][41]. The algal miRNA pathway is also distinct from that of higher plants, and the two evolutionary scenarios could explain those differences. The first scenario assumes that animal, higher plant, and algal miRNA pathways all evolved independently of each other. A second scenario takes into account that an animal-like miRNA pathway evolved early and persisted in lower plant lineages, including the green algae, though it was not retained in higher plants. The second scenario is more likely because C. reinhardtii and animal miRNA pathways share the presence of a Drosha-like structure (i.e., the presence of the proline-rich domain and the absence of the PAZ domain) of the miRNA processing enzyme [23].
Although evolutionary studies indicate that the ribonuclease Dicer is absent in bacteria and archaea, the helicase domain of Dicer displays high sequence similarity to its prokaryotic orthologues, archaeal Hef proteins that belong to Superfamily 2 (SF2) helicases [42][43]; the RNase III domains of Dicer are highly similar to bacterial RNase III [44]. A combination of the SF2 helicase and RNAse III domains in a single protein is considered a eukaryotic signature, and is supposed to be one of the critical, early events that lead to the consolidation of the eukaryotic RNA interference (RNAi) system [44].
3. Dicer Structure and the Importance of Its Domains
The hDicer ribonuclease is encoded by the DICER1 gene located on chromosome 14 in the subtelomeric region 14q32.13. The DICER1 gene spans a region of about 72 kbp and contains 29 exons. DICER1 is considered a housekeeping gene [45], although, compared to other housekeeping genes, it has an unusually long 3′-untranslated region (3′-UTR), i.e., over 4000 base pairs (bp) [45]. The protein product of DICER1 consists of 1922 amino acids (~220 kDa), and similar to other members of the third subfamily of RNases III, comprises a putative helicase domain, DUF283, PAZ, two RNase III domains (RNase IIIa and RNase IIIb) and dsRBD (Figure 1) [11]. Structural studies on hDicer have revealed new functional domains adjacent to the PAZ domain, namely, Platform and Connector helix [46][47]; the Platform-PAZ-Connector helix fragment is often referred to as “hDicer PAZ cassette” [47] or “PPC cassette” [48]. The experiments conducted with the use of cryo-electron microscopy (cryo-EM) techniques, revealed that the three-dimensional structure of the hDicer resembles the letter L [46][49]. Within this structure, the head, core and base can be distinguished. The head constitutes the PAZ and the Platform domains, the RNase III domains are in the core, while the helicase domain forms the base (Figure 2 and Figure 3A) [49].
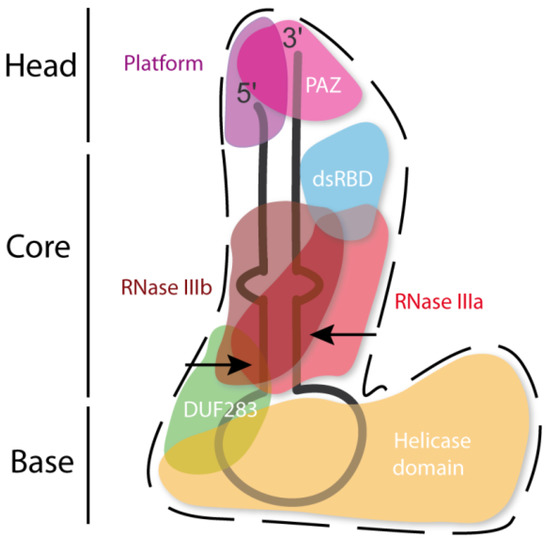
Figure 2. Scheme of the tertiary structure of hDicer in complex with pre-miRNA, prepared based on Liu et al. [49]. Arrows indicate the pre-miRNA cleavage sites by RNase III domains.
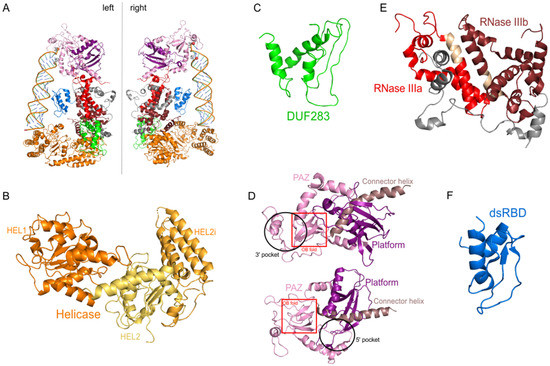
Figure 3. The 3D structure of hDicer (PDB entry 5ZAL) and hDicer individual domains visualized by PyMOL. (A) hDicer in complex with pre-let-7 (the loop region is not seen in this structure), the 5′-phosphate and the 3′-hydroxyl are indicated with spheres; (B) the helicase domain, with its three internal subdomains, is indicated in orange and yellow; (C) the DUF283 domain is indicated in green; (D) the Platform–PAZ–Connector helix cassette is shown in two orientations; the upper panel presents the 3′ pocket, whereas the lower panel exposes the 5′ pocket. The Platform domain is indicated in purple, PAZ in light pink, and Connector helix in brown. The OB fold within the PAZ domain is marked by the red rectangle; (E) the catalytic core—RNase IIIa and IIIb domains are indicated in red and wine red, respectively; fragments connecting RNase III domains are indicated in gray; fragments constituting the RNase III signature motifs are colored in beige; (F) the dsRBD domain is indicated in blue.
4. A Variety of Dicer Functions
In addition to miRNA and siRNA production, Dicer has been implicated in the biogenesis of a group of small RNAs, which are derived from tRNAs, called tRNA-derived fragments (tRFs) [50]. Studies on A. thaliana suggest that DCL proteins could be involved in tRF biogenesis in plants [51]. However, similar data from Drosophila and S. pombe revealed that the lack of Dicer does not significantly affect tRF levels [52]. Some tRNAs can fold into structures that resemble canonical Dicer substrates, thus it is possible that Dicer-dependent tRFs processing has evolved in certain organisms as a mechanism with biological relevance [53].
Beyond the biogenesis of small regulatory RNAs, Dicer also performs functions that are related to its nuclear localization. In organisms such as flies, plants and fission yeast, nuclear Dicer is involved in formation of heterochromatin [54]. In addition, it was demonstrated that, in mouse embryonic stem cells, chromatin modifications and DNA methylation of centromeric repeats are affected upon Dicer knockout, leading to accumulation of RNAs transcribed from centromeric repeats [55]. More recently, it was shown that, in mouse germ cells, Dicer is necessary for normal spermatogenesis, since during this process it controls pericentric heterochromatin expression [56]. Additionally, it was demonstrated in mice that Dicer is highly expressed in developing cerebrum, and lack of this protein causes the loss of cerebellar progenitor cells, presumably due to the accumulation of DNA damage that triggers apoptosis [53]. These results implicate Dicer involvement in DNA repair. Interestingly, it was shown in human and mouse cell lines, that inactivation of Dicer leads to inefficient DNA Damage Response (DDR) and the reduction in the level of Dicer-dependent small RNAs (ddRNAs), the regulatory RNAs involved in this process [57]. Another study reported that Dicer is involved in DNA double-strand break (DSB) repair [58], and DSB repair efficiency is reduced in A. thaliana in DCL2, DCL3 and DCL4 mutants [59]. Interestingly, it was demonstrated that hDicer is phosphorylated at position S1016 (the serine residue within the PPC cassette) upon induction of DSBs and recruited to facilitate DDR in the nucleus [60]. Nuclear Dicer was also implicated in the transcription-independent, global-genomic nucleotide excision repair pathway, in which it mediates the recruitment of a histone methyltransferase complex to the DNA damage site [61]. This way, Dicer presumably facilitates chromatin decondensation that, in consequence, allows repair factors to access the damage sites [62].
Dicer ribonuclease was found to be directly involved in the apoptosis process [63]. Apoptosis is a natural process of programmed cell death in multicellular organisms. One of the hallmarks of apoptosis is the fragmentation of chromosomal DNA leading to irreversible cell death. The process of apoptosis itself can be regulated by the Dicer-produced miRNAs [64]. Nevertheless, in a nematode C. elegans the apoptosis-induced caspases can cleave Dicer within the RNase IIIa domain, which results in releasing the C-terminal fragment of Dicer. Such truncated Dicer does not act as an RNase III enzyme; instead it displays the activity characteristic of DNases by initiating the chromosome fragmentation in the nucleus [63]. Such DNase activity has not been found for vertebrate Dicers; however, it was demonstrated that in human astrocytes, hypoxia-induced apoptosis is mediated by a decrease in hDicer levels via caspase-1 activation [65].
References
- Hansen, S.R.; Aderounmu, A.M.; Donelick, H.M.; Bass, B.L. Dicer’s Helicase Domain: A Meeting Place for Regulatory Proteins. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 185–193.
- Bass, B.L.; Hurst, S.R.; Singer, J.D. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 1994, 4, 301–314.
- Provost, P.; Samuelsson, B.; Rådmark, O. Interaction of 5-lipoxygenase with cellular proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 1881–1885.
- Matsuda, S.; Ichigotani, Y.; Okuda, T.; Irimura, T.; Nakatsugawa, S.; Hamaguchi, M. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta 2000, 1490, 163–169.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Jacobsen, S.E.; Running, M.P.; Meyerowitz, E.M. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 1999, 126, 5231–5243.
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626.
- Meyers, B.C.; Axtell, M.J. MicroRNAs in Plants: Key Findings from the Early Years. Plant Cell 2019, 31, 1206–1207.
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491.
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198.
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68.
- Elela, S.A.; Igel, H.; Ares, M. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 1996, 85, 115–124.
- Chanfreau, G.; Elela, S.A.; Ares, M.; Guthrie, C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997, 11, 2741–2751.
- Abou Elela, S.; Ares, M. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998, 17, 3738–3746.
- Chanfreau, G.; Rotondo, G.; Legrain, P.; Jacquier, A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998, 17, 3726–3737.
- Bardwell, J.C.; Régnier, P.; Chen, S.M.; Nakamura, Y.; Grunberg-Manago, M.; Court, D.L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989, 8, 3401–3407.
- Zer, C.; Chanfreau, G. Regulation and surveillance of normal and 3′-extended forms of the yeast aci-reductone dioxygenase mRNA by RNase III cleavage and exonucleolytic degradation. J. Biol. Chem. 2005, 280, 28997–29003.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Chong, M.M.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960.
- Wu, H.; Xu, H.; Miraglia, L.J.; Crooke, S.T. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000, 275, 36957–36965.
- Liang, X.H.; Crooke, S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011, 39, 4875–4889.
- Johanson, T.M.; Lew, A.M.; Chong, M.M. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013, 3, 130144.
- Valli, A.A.; Santos, B.A.; Hnatova, S.; Bassett, A.R.; Molnar, A.; Chung, B.Y.; Baulcombe, D.C. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016, 26, 519–529.
- Mukherjee, K.; Campos, H.; Kolaczkowski, B. Evolution of animal and plant dicers: Early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 2013, 30, 627–641.
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81.
- De Jong, D.; Eitel, M.; Jakob, W.; Osigus, H.J.; Hadrys, H.; Desalle, R.; Schierwater, B. Multiple dicer genes in the early-diverging metazoa. Mol. Biol. Evol. 2009, 26, 1333–1340.
- Kurzynska-Kokorniak, A.; Koralewska, N.; Pokornowska, M.; Urbanowicz, A.; Tworak, A.; Mickiewicz, A.; Figlerowicz, M. The many faces of Dicer: The complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res. 2015, 43, 4365–4380.
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011, 333, 1592.
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 2013, 9, e1003089.
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104.
- Bouché, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006, 25, 3347–3356.
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755.
- Chan, S.W.; Zilberman, D.; Xie, Z.; Johansen, L.K.; Carrington, J.C.; Jacobsen, S.E. RNA silencing genes control de novo DNA methylation. Science 2004, 303, 1336.
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221.
- Tworak, A.; Urbanowicz, A.; Podkowinski, J.; Kurzynska-Kokorniak, A.; Koralewska, N.; Figlerowicz, M. Six Medicago truncatula Dicer-like protein genes are expressed in plant cells and upregulated in nodules. Plant Cell Rep. 2016, 35, 1043–1052.
- Liu, Q.; Feng, Y.; Zhu, Z. Dicer-like (DCL) proteins in plants. Funct. Integr. Genom. 2009, 9, 277–286.
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250.
- Casas-Mollano, J.A.; Rohr, J.; Kim, E.J.; Balassa, E.; van Dijk, K.; Cerutti, H. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 2008, 179, 69–81.
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524.
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108.
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221.
- Aravind, L.; Walker, D.R.; Koonin, E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999, 27, 1223–1242.
- Lestini, R.; Delpech, F.; Myllykallio, H. DNA replication restart and cellular dynamics of Hef helicase/nuclease protein in Haloferax volcanii. Biochimie 2015, 118, 254–263.
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008, 23, 578–587.
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207.
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440.
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Park, J.E.; Heo, I.; Kim, V.N.; Patel, D.J. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616.
- Koralewska, N.; Ciechanowska, K.; Pokornowska, M.; Figlerowicz, M.; Kurzyńska-Kokorniak, A. Human ribonuclease Dicer—Structure and functions. Postepy Biochem. 2019, 65, 173–182.
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1549–1550.
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160.
- Alves, C.S.; Vicentini, R.; Duarte, G.T.; Pinoti, V.F.; Vincentz, M.; Nogueira, F.T. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol. Biol. 2017, 93, 35–48.
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78.
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986.
- Reyes-Turcu, F.E.; Grewal, S.I. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr. Opin. Genet. Dev. 2012, 22, 156–163.
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501.
- Yadav, R.P.; Mäkelä, J.A.; Hyssälä, H.; Cisneros-Montalvo, S.; Kotaja, N. DICER regulates the expression of major satellite repeat transcripts and meiotic chromosome segregation during spermatogenesis. Nucleic Acids Res. 2020, 48, 7135–7153.
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235.
- Chen, X.; Li, W.F.; Wu, X.; Zhang, H.C.; Chen, L.; Zhang, P.Y.; Liu, L.Y.; Ma, D.; Chen, T.; Zhou, L.; et al. Dicer regulates non-homologous end joining and is associated with chemosensitivity in colon cancer patients. Carcinogenesis 2017, 38, 873–882.
- Yao, Y.; Bilichak, A.; Golubov, A.; Kovalchuk, I. Arabidopsis thaliana siRNA biogenesis mutants have the lower frequency of homologous recombination. Plant Signal. Behav. 2016, 11, e1151599.
- Burger, K.; Schlackow, M.; Potts, M.; Hester, S.; Mohammed, S.; Gullerova, M. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J. Cell Biol. 2017, 216, 2373–2389.
- Chitale, S.; Richly, H. DICER- and MMSET-catalyzed H4K20me2 recruits the nucleotide excision repair factor XPA to DNA damage sites. J. Cell Biol. 2018, 217, 527–540.
- Chitale, S.; Richly, H. DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 2017, 45, 5901–5912.
- Nakagawa, A.; Shi, Y.; Kage-Nakadai, E.; Mitani, S.; Xue, D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 2010, 328, 327–334.
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154.
- Kang, Y.W.; Kim, Y.S.; Park, J.Y.; Chu, G.E.; Yang, Y.C.; Choi, B.Y.; Cho, W.G. Hypoxia-induced apoptosis of astrocytes is mediated by reduction of Dicer and activation of caspase-1. Cell Biol. Int. 2020, 44, 1394–1404.





