
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Aarti Krishnan | + 1768 word(s) | 1768 | 2021-02-08 07:52:41 | | | |
| 2 | Vicky Zhou | -2 word(s) | 1766 | 2021-02-12 10:28:51 | | | | |
| 3 | Vicky Zhou | -2 word(s) | 1766 | 2021-02-12 10:55:31 | | |
Video Upload Options
The review covers the current knowledge on the presence of amino acid biosynthesis and uptake within the Apicomplexa phylum, focusing on human-infecting pathogens: Toxoplasma gondii and Plasmodium falciparum. Given the complex multi-host lifecycle of these pathogens, we hypothesize that amino acids are made, rather than acquired, depending on the host niche. We summarize the stage-specificities of the biosynthesis enzymes, the role of amini acid transporters and the relevance of amino acids for parasite pathogenesis in vivo.
1. Introduction
Apicomplexan Parasites, such as Toxoplasma, Plasmodium, and Cryptosporidium spp., cause significant morbidity and mortality in humans and in livestock. Together, they lead to over half a million deaths each year and enormous economic burden [1][2]. The extraordinary ability of these parasites to invade, adapt, and persist in diverse host niches is multifactorial and depends on highly specialized pathogen-driven mechanisms which have evolved over a hundred million years [3][4]. Communication between a pathogen and its host therefore relies on a wide and dynamic array of molecular interactions, including exchanges of key metabolites. Although significant progress has been made in understanding host–pathogen interplay, a comprehensive and in-depth grasp of the metabolic capabilities of the various parasite developmental stages has not been achieved. Successful parasite replication is accomplished with the acquisition of essential nutrients from the host, which represents a vast repository of metabolites. However, the precise contributions of parasite de novo biosynthesis pathways versus acquisition are yet to be delineated. Recent studies further indicate that the nutritional availabilities and capabilities of the parasites vary between in vitro and in vivo conditions [5]. Therefore, to fully understand the underlying pathways, global, large-scale, and exhaustive experimental and computational approaches are becoming indispensable.
Indeed, in the last few years, our understanding of apicomplexan metabolism has improved with the assembly and curation of parasite genomes [6]. This information has enabled the generation of genome-scale metabolic networks in particular for T. gondii and Plasmodium spp. [5][7][8][9][10], which collectively encompass all known and experimentally validated genes encoding for proteins and enzymes associated with metabolic reactions. The biochemical reactions, localized to specific subcellular compartments, ultimately lead to the production of biomass, which constitutes the objective function of the mathematical framework. The framework can then be used to perform in silico simulations and to yield insights into gene and reaction essentialities, synthetic lethality, and in silico minimal media predictions. To challenge and improve the predictive power of these models, the latest reconstructions for T. gondii (iTgo) and the malaria parasites (P. falciparum and P. berghei (iPfa and iPbe, respectively)) were incorporated with valuable and recently obtained global experimental data. These datasets included large-scale genome-wide information on the fitness of parasites upon gene disruption [11][12][13], transcriptomics/RNAseq profiles of genes across lifecycle stages [14][15][16], and/or qualitative and quantitative metabolomics data [17][18][19]. Conceivably, incorporation of the different layers of –omics data (genomics, transcriptomics, and metabolomics) into the metabolic networks led to improved consistency with experimental data [5][10]. More recently, an impressive dataset assigned subcellular locations to over 2500 proteins in T. gondii using the hyperLOPIT (hyperplexed localization of organelle proteins by isotope tagging) method [20]. For a genome-scale model such as iTgo, precise cellular localization of enzymes enables accurate predictions on the feasibility of a reaction in a given compartment. Therefore, in an iterative cycle, the integration of high-throughput –omics data into computational frameworks will improve model-guided investigations for experimental studies, data which can be fed back into the network for another round of predictions.
In silico analysis on the curated and harmonized iTgo model revealed that pathways for the de novo biosynthesis of several amino acids was dispensable [5]. This led us to hypothesize that, in the fast-replicating stage of the parasite (hereafter referred to as tachyzoites), most of the amino acids were acquired from the host. Dedicated transport mechanisms for some of the amino acids have been investigated [21][22] and uptake of proteins and subsequent proteolytic degradation within a specialized vacuolar organelle (VAC) has previously been described [23]. However, transporters for several other amino acids are still unidentified, opening numerous doors for future investigation. Interestingly, the enzymes that do exist for de novo synthesis or interconversion of amino acids are transcriptionally upregulated in other developmental stages of the parasites (slow-growing bradyzoites, merozoites, gametes, or oocysts) compared to tachyzoites [14].
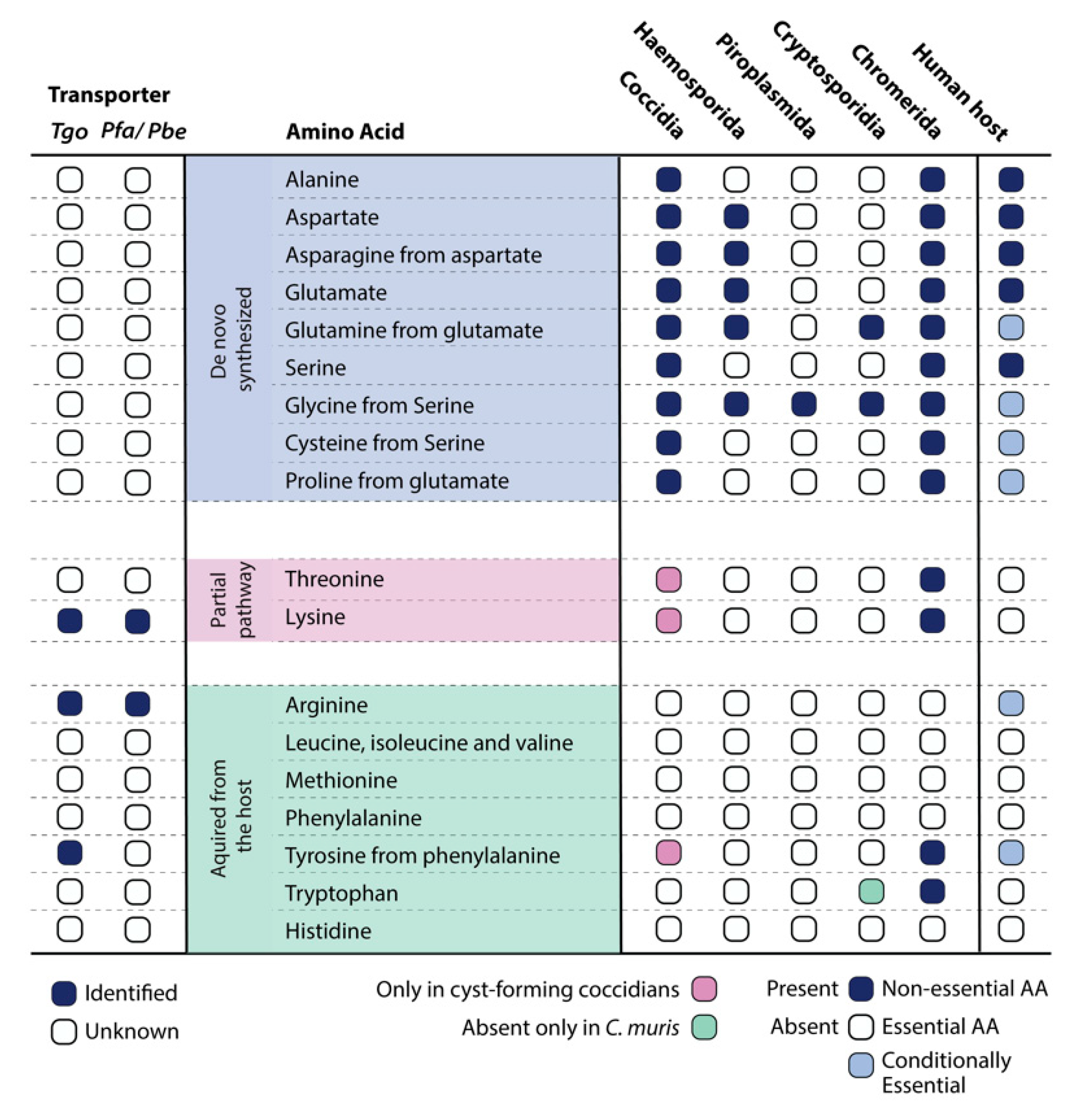
Genomic analysis further illustrates that, among the Apicomplexa, the cyst-forming coccidian subgroup along with Chromeridae, which shares a proto-apicomplexan ancestor, have the greatest number of enzymes for the synthesis of amino acids (Figure 1). As expected, in certain genera such as the Cryptosporidia and Piroplasmida, a large number of genes have been lost. This is likely reflective of their intracellular lifestyle, as Cryptosporidia develop in an extra-cytoplasmic compartment on the apical surface of intestinal epithelium cells and piroplasms proliferate freely within the host cytosol, enabling direct access to nutrients from the gut microbiome and host cell, respectively [24][25][26][27].
2. Amino Acid Metabolism in Apicomplexan Parasites
Amino acids are vital to survival, not only to make up proteins but also as key molecules for the proper functioning of metabolic pathways. Recent technologies have enabled us to understand the importance of the biosynthesis versus acquisition routes of amino acids in human-infecting pathogens. A combination of experimental and computational approaches, such as genome-wide metabolic models, curated and harmonized with CRISPR-Cas9 screening, transcriptomics, and metabolomics data, have been instrumental in deepening our understanding of the underlying mechanisms. Using our genome-scale metabolic model for the fast-replicating stage of T. gondii, iTgo, we analyzed the pathways for the biosynthesis of amino acids. A comparison of our model predictions with experimental CRISPR-Cas9 screening data revealed that enzymes to produce amino acids in the tachyzoite stage were dispensable, leading us to hypothesize the utilization of these enzymes in a different developmental stage [5]. This observation was indeed supported by the increased transcript levels of the biosynthesis pathways in the bradyzoite, merozoite, gamete, and oocyst stages of T. gondii [14].
Thorough investigations of each of the 20 amino acids disclosed the existence of two (partial) biosynthesis pathways for threonine and lysine within the parasite, but they were found missing in the human host. This highlights potential opportunities for drug discovery and development, specifically targeting the parasite’s metabolism. Moreover, by comparing the genomes of species within the Apicomplexa phylum, the non-cyst forming branch (Eimeria and Cyclospora spp.) lacks the enzymes for threonine and lysine biosynthesis (Figure 1). It likely indicates the need for de novo synthesis of these amino acids during encystation and calls for future work to identify the missing enzymes and their importance during latency or a different developmental stage. Fascinatingly and unique to a few apicomplexans, Cardiosporidium cionae appears to have maintained a bacterial endosymbiont that contains the pathway for de novo lysine biosynthesis [28]. It provides only two key metabolites: lysine and lipoic acid, a mechanism that cleverly reduces host dependency and potential virulence, making the extracellular life of C. cionae possible [28].
Although capable of synthesizing quite a few amino acids (alanine, aspartate, asparagine, glutamine, glutamate, glycine, cysteine, and proline), apicomplexan parasites, at least in their fast-replicating stages, primarily rely on uptake from the host. Especially during the intraerythrocytic stage, the amino acids requirement of the malaria parasites is largely met via host-derived hemoglobin degradation within a digestive vacuole (DV) [29][30]. Several aminopeptidases are known to be harbored within the DV, where breakdown proteins generate short peptides and amino acids, which can be transported out of the DV [31][32]. Given such a major reliance on the host for amino acids, identifying the transporter proteins and other amino acid acquisition mechanisms will prove to be of significant interest for drug targeting. According to Parker et al., the transmembrane transporters identified in T. gondii share the classical domains but are largely unspecific (transporting more than one amino acid) [21]. The study included 16 transporter-like proteins; however, experimental data suggested that only a few displayed a fitness-conferring phenotype [21]. The dispensable transporters therefore represent potential candidates for which the parasite has biosynthesis capabilities or redundant transport mechanisms. Performing high-throughput functional screens to identify synthetic lethality between the known enzymes and/or transporters could hence yield valuable information.
Most apicomplexans possess two endosymbionts: a unique relic of a plastid organelle called the apicoplast, that resulted from an ancient secondary endosymbiosis event and a single tubular mitochondrion. Both of these organelles harbor several essential metabolic pathways and utilize amino acids for their proper functioning. In order to fulfill the translational requirements of these organelles, the parasite must maintain a supply of charged tRNAs. Aminoacyl-tRNA synthetases (aaRSs) play an essential role in this process by charging the tRNAs with their cognate amino acids. Interestingly, in T. gondii, the apicoplast translation relies on its own set of tRNAs and imported amino acids and aaRSs, while the mitochondrion exclusively depends on the cytosolic components [33]. Given the essentiality of the aaRSs for ribosomal protein synthesis, several of them have been suggested and characterised as promising antiparasitic targets [34][35][36]. Inhibiting the transport of the amino acids and charged tRNAs to the endosymbiotic compartments could further present novel drug targets for intervention.
As highlighted, it is well known that amino acids are essential for protein production, but more recently, some light has been shed on the interactions between amino acid metabolism and modulation of the host’s immune response. In vivo studies in mice showed that supplementation of a cocktail of amino acids (arginine, lysine, and valine (RKV)) during the liver stage of Plasmodium parasites abolished proliferation, as the amino acids played a crucial role in boosting the innate immune response. RKV activated MyD88, a crucial adaptor molecule involved in Toll-like receptor (TLR)-mediated signal transduction, essential for the function of myeloid cells and subsequent parasite elimination [37]. In parallel studies on cerebral malaria, the most severe symptom of malaria, where parasites destroy the blood–brain barrier, arginine supplementation was shown to increase cerebral blood flow, thereby partially reversing the several damages caused by cerebral ischemia [38]. These observations warrant further exploration of metabolite-based strategies against the apicomplexans and focus on the interface between amino acid metabolism and eradication of parasite burden.
Lastly, little knowledge has been gained on the status of amino acids in the chronic, slow-growing and dormant stages of both T. gondii and Plasmodium hypnozoites. Therefore, future work must focus on constructing in silico stage-specific metabolic networks and on generating experimental data to shed light on these enigmatic stages. Targeting either the biosynthesis capabilities or acquisition pathways or both would enable better intervention strategies that could put an end not only to parasite proliferation but also to persistence and transmission of these disease-causing pathogens.
References
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923.
- Torgerson, P.R.; Mastroiacovo, P. The global burden of congenital toxoplasmosis: A systematic review. Bull. World Health Organ. 2013, 91, 501–508.
- Escalante, A.A.; Ayala, F.J. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc. Natl. Acad. Sci. USA 1995, 92, 5793–5797.
- White, M.W.; Suvorova, E.S. Apicomplexa Cell Cycles: Something Old, Borrowed, Lost, and New. Trends Parasitol. 2018, 34, 759–771.
- Krishnan, A.; Kloehn, J.; Lunghi, M.; Chiappino-Pepe, A.; Waldman, B.S.; Nicolas, D.; Varesio, E.; Hehl, A.; Lourido, S.; Hatzimanikatis, V.; et al. Functional and Computational Genomics Reveal Unprecedented Flexibility in Stage-Specific Toxoplasma Metabolism. Cell Host Microbe 2020, 27, 290–306.e11.
- Aurrecoechea, C.; Barreto, A.; Basenko, E.Y.; Brestelli, J.; Brunk, B.P.; Cade, S.; Crouch, K.; Doherty, R.; Falke, D.; Fischer, S.; et al. EuPathDB: The eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017, 45, D581–D591.
- Song, C.; Chiasson, M.A.; Nursimulu, N.; Hung, S.S.; Wasmuth, J.; Grigg, M.E.; Parkinson, J. Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii. Mol. Syst. Biol. 2013, 9, 708.
- Chiappino-Pepe, A.; Tymoshenko, S.; Ataman, M.; Soldati-Favre, D.; Hatzimanikatis, V. Bioenergetics-based modeling of Plasmodium falciparum metabolism reveals its essential genes, nutritional requirements, and thermodynamic bottlenecks. PLoS Comput. Biol. 2017, 13, e1005397.
- Tymoshenko, S.; Oppenheim, R.D.; Agren, R.; Nielsen, J.; Soldati-Favre, D.; Hatzimanikatis, V. Metabolic Needs and Capabilities of Toxoplasma gondii through Combined Computational and Experimental Analysis. PLoS Comput. Biol. 2015, 11, e1004261.
- Stanway, R.R.; Bushell, E.; Chiappino-Pepe, A.; Roques, M.; Soldati-Favre, D.; Janse, C.J.; Hatzimanikatis, V.; Billker, O.; Heussler, V.T. Genome Scale Identification of Essential Metabolic Processes for Targeting the Plasmodium Liver Stage. Cell 2019, 179, 1112–1128.
- Zhang, M.; Wang, C.; Otto, T.D.; Oberstaller, J.; Liao, X.; Adapa, S.R.; Udenze, K.; Bronner, I.F.; Casandra, D.; Mayho, M.; et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 2018, 360, eaap7847.
- Bushell, E.; Gomes, A.R.; Sanderson, T.; Anar, B.; Girling, G.; Herd, C.; Metcalf, T.; Modrzynska, K.; Schwach, F.; Martin, R.E.; et al. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 2017, 170, 260–272.e8.
- Sidik, S.M.; Huet, D.; Ganesan, S.M.; Huynh, M.-H.H.; Wang, T.; Nasamu, A.S.; Thiru, P.; Saeij, J.P.J.; Carruthers, V.B.; Niles, J.C.; et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 2016, 166, 1423–1435.e12.
- Hehl, A.B.; Basso, W.U.; Lippuner, C.; Ramakrishnan, C.; Okoniewski, M.; Walker, R.A.; Grigg, M.E.; Smith, N.C.; Deplazes, P. Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genom. 2015, 16, 66.
- Otto, T.D.; Böhme, U.; Jackson, A.P.; Hunt, M.; Franke-Fayard, B.; Hoeijmakers, W.A.M.; Religa, A.A.; Robertson, L.; Sanders, M.; Ogun, S.A.; et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014, 12, 86.
- Caldelari, R.; Dogga, S.; Schmid, M.W.; Franke-Fayard, B.; Janse, C.J.; Soldati-Favre, D.; Heussler, V. Transcriptome analysis of Plasmodium berghei during exo-erythrocytic development. Malar. J. 2019, 18, 330.
- Teng, R.; Junankar, P.R.; Bubb, W.A.; Rae, C.; Mercier, P.; Kirk, K. Metabolite profiling of the intraerythrocytic malaria parasite Plasmodium falciparum by 1H NMR spectroscopy. NMR Biomed. 2009, 22, 292–302.
- Teng, R.; Lehane, A.M.; Winterberg, M.; Shafik, S.H.; Summers, R.L.; Martin, R.E.; Van Schalkwyk, D.A.; Junankar, P.R.; Kirk, K. 1H-NMR metabolite profiles of different strains of Plasmodium falciparum. Biosci. Rep. 2014, 34, 685–699.
- Vo Duy, S.; Besteiro, S.; Berry, L.; Perigaud, C.; Bressolle, F.; Vial, H.J.; Lefebvre-Tournier, I. A quantitative liquid chromatography tandem mass spectrometry method for metabolomic analysis of Plasmodium falciparum lipid related metabolites. Anal. Chim. Acta 2012, 739, 47–55.
- Barylyuk, K.; Koreny, L.; Ke, H.; Butterworth, S.; Crook, O.M.; Lassadi, I.; Gupta, V.; Tromer, E.; Mourier, T.; Stevens, T.J.; et al. A Comprehensive Subcellular Atlas of the Toxoplasma Proteome via hyperLOPIT Provides Spatial Context for Protein Functions. Cell Host Microbe 2020, 28, 752–766.e9.
- Parker, K.E.R.; Fairweather, S.J.; Rajendran, E.; Blume, M.; McConville, M.J.; Bröer, S.; Kirk, K.; van Dooren, G.G. The tyrosine transporter of Toxoplasma gondii is a member of the newly defined apicomplexan amino acid transporter (ApiAT) family. PLoS Pathog. 2019, 15, e1007577.
- Rajendran, E.; Hapuarachchi, S.V.; Miller, C.M.; Fairweather, S.J.; Cai, Y.; Smith, N.C.; Cockburn, I.A.; Broër, S.; Kirk, K.; Van Dooren, G.G. Cationic amino acid transporters play key roles in the survival and transmission of apicomplexan parasites. Nat. Commun. 2017, 8, 1–13.
- Dou, Z.; McGovern, O.L.; Di Cristina, M.; Carruthers, V.B. Toxoplasma gondii Ingests and Digests Host Cytosolic Proteins. MBio 2014, 5, e01188-14.
- Burgess, S.L.; Gilchrist, C.A.; Lynn, T.C.; Petri, W.A. Parasitic Protozoa and Interactions with the Host Intestinal Microbiota. Infect. Immun. 2017, 85, e00101-17.
- O’Hara, S.P.; Chen, X.-M. The cell biology of cryptosporidium infection. Microbes Infect. 2011, 13, 721–730.
- Mehlhorn, H.; Schein, E. The Piroplasms: Life Cycle and Sexual Stages. Adv. Parasitol. 1985, 23, 37–103.
- Jalovecka, M.; Hajdusek, O.; Sojka, D.; Kopacek, P.; Malandrin, L. The Complexity of Piroplasms Life Cycles. Front. Cell. Infect. Microbiol. 2018, 8, 248.
- Hunter, E.S.; Paight, C.; Lane, C.E. Metabolic Contributions of an Alphaproteobacterial Endosymbiont in the Apicomplexan Cardiosporidium cionae. Front. Microbiol. 2020, 11, 3014.
- Babbitt, S.E.; Altenhofen, L.; Cobbold, S.A.; Istvan, E.S.; Fennellf, C.; Doerig, C.; Llinás, M.; Goldberg, D.E. Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proc. Natl. Acad. Sci. USA 2012, 109, E3278–E3287.
- Goldberg, D.E. Complex nature of malaria parasite hemoglobin degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 5283–5284.
- Gavigan, C.S.; Dalton, J.P.; Bell, A. The role of aminopeptidases in haemoglobin degradation in Plasmodium falciparum-infected erythrocytes. Mol. Biochem. Parasitol. 2001, 117, 37–48.
- Lee, A.H.; Dhingra, S.K.; Lewis, I.A.; Singh, M.K.; Siriwardana, A.; Dalal, S.; Rubiano, K.; Klein, M.S.; Baska, K.S.; Krishna, S.; et al. Evidence for Regulation of Hemoglobin Metabolism and Intracellular Ionic Flux by the Plasmodium falciparum Chloroquine Resistance Transporter. Sci. Rep. 2018, 8, 1–13.
- Pino, P.; Aeby, E.; Foth, B.J.; Sheiner, L.; Soldati, T.; Schneider, A.; Soldati-Favre, D. Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNAMet formylation in Apicomplexa. Mol. Microbiol. 2010, 76, 706–718.
- Jain, V.; Yogavel, M.; Kikuchi, H.; Oshima, Y.; Hariguchi, N.; Matsumoto, M.; Goel, P.; Touquet, B.; Jumani, R.S.; Tacchini-Cottier, F.; et al. Targeting prolyl-tRNA synthetase to accelerate drug discovery against malaria, leishmaniasis, toxoplasmosis, cryptosporidiosis, and coccidiosis. Structure 2017, 25, 1495–1505.
- Palencia, A.; Liu, R.J.; Lukarska, M.; Gut, J.; Bougdour, A.; Touquet, B.; Wang, E.-D.; Li, X.; Alley, M.R.K.; Freund, Y.R.; et al. Cryptosporidium and Toxoplasma parasites are inhibited by a benzoxaborole targeting leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 2016, 60, 5817–5827.
- Cowell, A.N.; Winzeler, E.A. Advances in omics-based methods to identify novel targets for malaria and other parasitic protozoan infections. Genome Med. 2019, 11, 1–17.
- Meireles, P.; Brás, D.; Fontinha, D.; Chora, Â.F.; Serre, K.; Mendes, A.M.; Prudêncio, M. Elimination of Hepatic Rodent Plasmodium Parasites by Amino Acid Supplementation. iScience 2020, 23, 101781.
- Moreira, A.S.; Estato, V.; Malvar, D.C.; Sanches, G.S.; Gomes, F.; Tibirica, E.; Daniel-Ribeiro, C.T.; Carvalho, L.J.M. L-arginine supplementation and thromboxane synthase inhibition increases cerebral blood flow in experimental cerebral malaria. Sci. Rep. 2019, 9, 13621.




