
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Michail Kotsyfakis | + 2150 word(s) | 2150 | 2021-02-10 05:23:57 | | | |
| 2 | Catherine Yang | Meta information modification | 2150 | 2021-02-10 07:39:28 | | |
Video Upload Options
Protease inhibitors (PIs) are ubiquitous regulatory proteins present in all kingdoms. They play crucial tasks in controlling biological processes directed by proteases which, if not tightly regulated, can damage the host organism. PIs can be classified according to their targeted proteases or their mechanism of action.
1. Introduction
Proteases are ubiquitous enzymes in plants, animals, and microorganisms that play key roles in the majority of physiological processes [1][2]. Proteases are involved in several reversible and irreversible reaction cascades including hormone production/signaling pathways, apoptosis, inflammatory reactions, and homeostasis [3]. Depending on the active amino acid in the enzyme’s active site, proteases are classified into cysteine, serine, aspartic, glutamic, and threonine proteases [3], and metalloproteases represent another protease class containing a divalent metal ion linked to the active site residues [4]. Serine proteases are the most abundant proteolytic enzymes, followed by metallo- and cysteine proteases and finally the aspartic and threonine proteases [5]. In addition to their roles in vital biochemical processes, proteases are also implicated in various diseases such as viral diseases, cancer, inflammation, and bleeding disorders [6]. Given their important roles in diverse physiological processes, protease activity must be rigorously controlled and regulated to avoid any enzyme dysregulation that might be pathogenic to the host organism [7]. This tight regulation is usually conducted by blocking the zymogen, the inactive enzyme precursor, or through the action of protease inhibitors (PIs) [8], which partially or totally inhibit enzymes by forming a complex with their target proteases [4].
While the majority of PIs are proteins or peptides, some low molecular weight non-proteinaceous compounds such as polysaccharides, glycerolipids, triterpenes, and polyphenols are also considered PIs [9]; these are not specific to particular proteases and inhibit a broad spectrum of enzymes [10]. In contrast, proteinaceous PIs are usually more specific and can even target unique proteases. As a result, PIs can be classified according to their target enzyme, although exceptions are frequently encountered, such as the α2-macroglobulin that inhibits proteases of different classes [6]. A particularly impressive role for PIs has been observed in parasite–host crosstalk; PIs found in tick saliva have been shown to modulate host immune cells [11], mediating local immunosuppression and modulating blood clotting at the site of infection, thereby exerting a beneficial effect to the tick (allowing attachment and feeding) at the expense of the host [12].
Given their fundamental roles and potential translational application, there have been significant efforts to identify new PIs from various sources and study those already identified in detail using novel technologies and methods. Indeed, with technological advances, the study of PIs has substantially improved over recent years, not least the availability of three-dimensional (3D) structural information for several PIs and their targeted proteases, permitting receptor-based design.
Here, we review the biochemistry and fundamental mechanisms of action of PIs. We enumerate and discuss the different classes of PIs based on the proteases they inhibit and their mechanism of action. Moreover, we discuss their applications in critical fields like agriculture and medicine. In the final section, we focus on an interesting natural source of PIs, tick salivary glands, and their potential pharmacological applications.
2. Classification of Protease Inhibitors
The catalytic activity of proteases is regulated by different inhibition mechanisms and different PI families [13]. Despite similarities in the 3D structure of PIs, they can be classified into over 107 families and divided into 40 clans according to their structural similarities (secondary and tertiary) and their different functions [13]. Laskowski and Kato first developed a classification scheme for PIs in 1980 [14] according to their reactive center, disulfide bond number, and amino acid sequence [14]. With advances in biotechnology and increasing knowledge about PIs, Rawlings et al. [15] established a classification of PIs in 2004 based on amino acid sequence homology that subclassified PIs to 48 families and 26 clans [5].
2.1. Target-Based Classification
PIs can be classified according to their target protease into six groups [4]: serine, cysteine, aspartyl, glutamate, and threonine protease inhibitors. Metalloprotease inhibitors are also contained within this classification, as they inhibit proteases with a divalent metal ion in their active site [16]. A non-exhaustive list of the most common PI families, their principal features, and their properties according to the target-based classification system is illustrated in Figure 1 and discussed below.
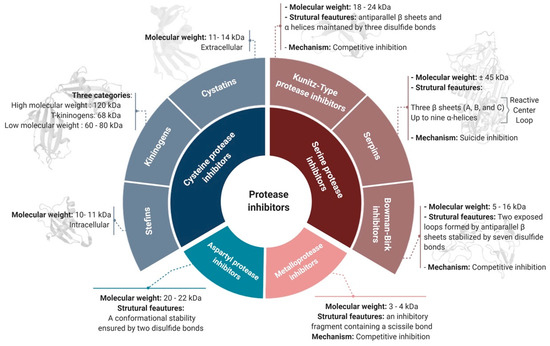
Figure 1. The target-based classification of protease inhibitors.
2.1.1. Serine Protease Inhibitors
Since serine proteases are the most abundant protease family, containing >26,000 proteases [17], their respective inhibitors are the largest group of PIs in animals, plants, and microorganisms [18]. Most serine protease inhibitors follow the conventional mechanism of inhibition through the generation of irreversible Michaelis complexes characterized by covalent bonds between the protease and the inhibitor [19]. Serine PI domains ensure efficient functionality and allow their subclassification into various superfamilies such as the Kunitz-type PIs, Bowman–Birk inhibitors, serpins, trypsin inhibitor-like domain inhibitors (TILs), and Kazal domain inhibitors [12].
Serpins
The serpin superfamily is the largest serine PI family [20]. Serpins typically weigh ~45 kDa and are relatively large molecules (~350–400 amino acids) compared to other PIs [21]. The 3D structure of serpins includes three β-sheets (A, B, and C) and up to nine α-helices that fold to form a specific spatial structure including a reactive center loop (RCL) near the C-terminus [22]. Serpins are categorized as “suicide inhibitors” due to the inactive covalent complex formed with their target protease [23]. This suicide inhibition is often referred to as a “mousetrap”, since the RCL interacts with the target protease active site, and its scissile bond (P1-P1’) is cleaved to generate a stable complex [24]. The resulting bond between the protease and the RCL leads to the insertion of the cleaved RCL into the β-sheet A and relocation of the protease to the opposite pole of the serpin, forming a suicide covalent complex [24]. Serpins are involved in the regulation of different physiological processes such as blood coagulation, fibrinolysis, signal transduction, the complement cascade, and immune responses [25].
Kunitz-Type PIs
PIs in the Kunitz superfamily are characterized by the presence of one or many Kunitz inhibitory domains. They are generally small proteins with molecular weights ranging from 18 to 24 kDa [26]. The Kunitz domain is characterized by anti-parallel β-sheets and α-helices maintained in a compact 3D structure by three disulfide bonds [27]. Most Kunitz-type PIs are competitive inhibitors, acting in a substrate-like manner and binding reversibly to the protease [28]. Active site blocking is mediated by the RCL attachment to the catalytic zone through a non-covalent bond. The highly exposed RCL loop of Kunitz-domain inhibitors is suitable for a wide variety of proteases, so these inhibitors are relatively non-specific and therefore potentially useful across a range of applications [29]. Indeed, Kunitz-type PIs are known to regulate inflammation and coagulation factors and have also been implicated in tumor biology [3].
Bowman–Birk Inhibitors (BBIs)
This superfamily of PIs is characterized by small molecular weight peptides ranging from 5 to 16 kDa and a structure with a single or two inhibitory regions [30]. BBIs are competitive inhibitors and follow the classical mechanism of substrate binding to the protease active site [31]. A single BBI inhibitor protein can act on two different target proteases simultaneously by virtue of two opposed loops formed by antiparallel β-sheets and stabilized by seven disulfide bonds [23][32]. Given their specific mechanism, several researchers have focused on BBIs for specific applications such as to inhibit cancer [33].
2.1.2. Cysteine Protease Inhibitors
Cysteine protease inhibitors (CPIs), or cystatins, are the second largest group of PIs after serine PIs. They are divided into three main families: family-1 cystatins or stefins, family-2 cystatins or cystatins, and family-3 cystatins or kininogens. Stefins are mostly intracellular and the smallest cystatin family in terms of molecular weight (10–11 kDa) [3]. Stefins inhibit cathepsins B, L, and H and also papain. In several therapeutic investigations, they have been identified as potential diagnostic tools for cancer [34]. Like stefins, cystatins inhibit papain and cathepsins B, L, and H, but, they are larger (11–14 kDa) and are transported out of the cell to exert their action [35]. Kininogens are divided into three categories: high molecular weight kininogens (120 kDa), T-kininogens (68 kDa), and low molecular weight kininogens (60–80 kDa) [3][36][37]. They play important roles in the modulation of inflammatory responses and are used as biomarkers of kidney disorders [37].
There are numerous documented functions of CPIs, and some have been shown to be critical for the proper functioning of important physiological pathways such as cathepsin regulation [38]. The structural features of CPIs include 4 to 5 antiparallel β-sheets surrounding an α-helix. Their highly conserved inhibitory domain is mainly composed of two hairpin-like loops formed by the β-sheets and the N-terminal region [39]. Cystatins follow the competitive inhibition model with slight modifications, as they do not bind in a substrate-like manner. In fact, the two hairpin loops bind to the protease active site and block the access of any substrate, while the N-terminal region maintains effective attachment of the inhibitor to the enzyme [40][41].
2.1.3. Metalloproteases Inhibitors
Despite their low molecular weight (only 3–4 kDa), metalloprotease inhibitors (MPIs) effectively inhibit a wide range of metalloproteases [42]. MPIs are classified as competitive inhibitors, since they act in a substrate-like manner [43]. From a structural point of view, MPIs do not possess an inhibitory loop or a specific secondary structure for inhibition. Instead, their inhibitory fragment is located near the C-terminus and contains a scissile bond. The cleavage of this latter bond allows the fixation of the new C-terminal side to the active site of the protease with the help of its metallic ion [23][28]. The efficiency of MPI inhibition has been reported to be enhanced by secondary interactions outside the active site of the protease [44].
2.1.4. Aspartyl Protease Inhibitors
The natural aspartyl PIs are proteins of about 20 to 22 kDa and conformational stability ensured by two disulfide bonds [45]. Despite aspartyl proteases including important members such as cathepsins D and E, renin, pepsin A and C, and, most importantly, the HIV-1 protease, their natural inhibitors remain poorly described for several reasons. One main reason is probably related to the low representation of aspartyl proteases in the human genome, with only 15 members described [46]. Regardless of their low bioavailability, the presence of a scissile bond and their short half-life have meant that strategies to inhibit aspartyl proteases involve the development of synthetic peptides or mimics with a non-cleavable bond to replace the scissile bond [47].
2.2. Mechanism-Based Classification
The target-based classification is limited, as numerous PIs are active against two or more enzymes. Indeed, in humans, there are vastly more proteases than PIs; despite the continuous discovery of human proteases and their respective inhibitors, the ratio of one PI for every five proteases has remained constant [41][48]. However, in addition to the target-based classification, it is possible to classify PIs according to their mechanism of inhibition, in particular the steric or catalytic inhibition of the enzyme active site or its neighboring regions [49]. Enzyme inhibition mechanisms can be divided into two general categories, reversible and irreversible. Reversible inhibition can be further subdivided into competitive, uncompetitive, and non-competitive inhibition [41]. The mechanism based-classification can be divided into three major classes (Figure 2): competitive protease inhibition (also called canonical inhibition), exosite-assisted competitive inhibition or non-canonical inhibition, and finally irreversible inhibition or trapping inhibition [41].
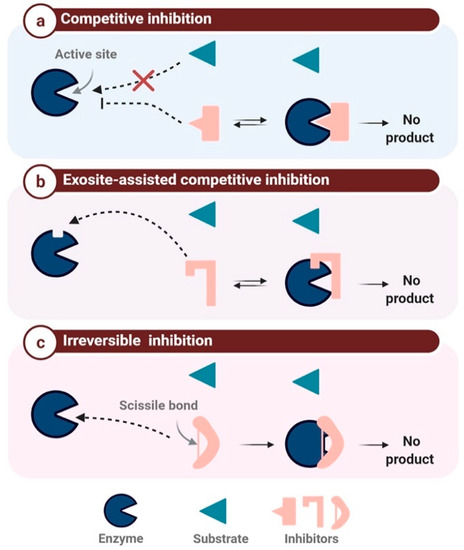
Figure 2. The mechanism-based classification of protease inhibitors. (a): competitive inhibition: the inhibitor binds to the active site instead of the substrate, (b): Exosite-assisted competitive inhibition: the inhibitor blocks the access to the active site through binding to an exosite, (c): Irreversible inhibition: the inhibitor binds irreversibly to the enzyme inducing its inactivation
2.2.1. Competitive Protease Inhibitors
The canonical inhibition mechanism is also known as the standard or Laskowski mechanism [50]. Inhibitors belonging to this class attach using a lock-and-key system through the insertion of the inhibitor RCL into the catalytic site of the targeted protease (Figure 2a). The β-sheet conformation allows the binding of the inhibitor to the active site in a substrate-like manner. Consequently, the RCL scissile bond is slowly hydrolyzed by the protease without any product release, as the amide bond is later reconnected [15][41]. Numerous PIs belong to this family, mainly the BBIs and Kazal and Kunitz domain-containing inhibitors.
2.2.2. Exosite-Assisted Competitive Inhibitors
Known also as non-canonical inhibitors, this class represents inhibitors binding to a secondary site distinct from the protease active site (Figure 2b). Access to this active site is blocked in a non-catalytic manner [51][52]. This inhibition mechanism is classified as competitive but differs from the standard Laskowski mechanism [50]. Inhibitor binding to the exosite is crucial for inhibition, as it maintains the inhibitor–enzyme interaction and enhances inhibitor specificity [51]. As mentioned above, CPIs are non-canonical PIs that follow a slightly modified competitive inhibition mechanism, since the N-terminal region does not interact with the active site in a substrate-like manner but switches to the side of the active site to ensure sufficient binding energy for the enzyme–inhibitor complex [41][53].
2.2.3. Irreversible Inhibition
This class of inhibition is triggered by the protease, which catalyzes the activation of its respective inhibitor (also referred to as a suicide substrate). The cleavage of the inhibitor reactive loops (Figure 2c) triggers a major conformational change, resulting in the irreversible cross-linking of the protease to its inhibitor [23][51]. α-2-macroglobulin is a 600 kDa inhibitor with four reactive loops on its surface, which plays a major role in the elimination of excessive proteases in the blood [54]. Serpins are also well-known suicide inhibitors, as described above [55].
References
- Hartl, M.; Giri, A.P.; Kaur, H.; Baldwin, I.T. The multiple functions of plant serine protease inhibitors: Defense against herbivores and beyond. Plant Signal. Behav. 2011, 6, 1009–1011.
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147.
- Shamsi, T.N.; Parveen, R.; Fatima, S. Characterization, biomedical and agricultural applications of protease inhibitors: A review. Int. J. Biol. Macromol. 2016, 91, 1120–1133.
- Harish, B.S.; Uppuluri, K.B. Microbial serine protease inhibitors and their therapeutic applications. Int. J. Biol. Macromol. 2018, 107, 1373–1387.
- Dunaevsky, Y.E.; Popova, V.V.; Semenova, T.A.; Beliakova, G.A.; Belozersky, M.A. Fungal inhibitors of proteolytic enzymes: Classification, properties, possible biological roles, and perspectives for practical use. Biochimie 2014, 101, 10–20.
- Sabotic, J.; Kos, J. Microbial and fungal protease inhibitors—Current and potential applications. Appl. Microbiol. Biotechnol. 2012, 93, 1351–1375.
- Oliva, M.L.; Sampaio, M.U. Action of plant proteinase inhibitors on enzymes of physiopathological importance. An. Acad. Bras. Cienc. 2009, 81, 615–621.
- Gagaoua, M.; Hafid, K.; Boudida, Y.; Becila, S.; Ouali, A.; Picard, B.; Boudjellal, A.; Sentandreu, M.A. Caspases and Thrombin Activity Regulation by Specific Serpin Inhibitors in Bovine Skeletal Muscle. Appl. Biochem. Biotechnol. 2015, 177, 279–303.
- Doljak, B.; Cateni, F.; Anderluh, M.; Procida, G.; Zilic, J.; Zacchigna, M. Glycerolipids as selective thrombin inhibitors from the fungus Stereum hirsutum. Drug Dev. Ind. Pharm. 2006, 32, 635–643.
- Overall, C.M.; Blobel, C.P. In search of partners: Linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 2007, 8, 245–257.
- Stibraniova, I.; Bartikova, P.; Holikova, V.; Kazimirova, M. Deciphering Biological Processes at the Tick-Host Interface Opens New Strategies for Treatment of Human Diseases. Front. Physiol. 2019, 10.
- Chmelar, J.; Kotal, J.; Langhansova, H.; Kotsyfakis, M. Protease Inhibitors in Tick Saliva: The Role of Serpins and Cystatins in Tick-host-Pathogen Interaction. Front. Cell. Infect. Microbiol. 2017, 7, 216.
- Rawlings, N.D.; Alan, J.; Thomas, P.D.; Huang, X.D.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632.
- Laskowski, M.; Kato, I. Protein Inhibitors of Proteinases. Annu. Rev. Biochem. 1980, 49, 593–626.
- Rawlings, N.D.; Tolle, D.P.; Barrett, A.J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004, 378, 705–716.
- Jacobson, A.R.; Adler, M.; Silvaggi, N.R.; Allen, K.N.; Smith, G.M.; Fredenburg, R.A.; Stein, R.L.; Park, J.B.; Feng, X.C.; Shoemaker, C.B.; et al. Small molecule metalloprotease inhibitor with in vitro, ex vivo and in vivo efficacy against botulinum neurotoxin serotype A. Toxicon Off. J. Int. Soc. Toxinol. 2017, 137, 36–47.
- Page, M.J.; Di Cera, E. Serine peptidases: Classification, structure and function. Cell. Mol. Life Sci. 2008, 65, 1220–1236.
- Billinger, E.; Zuo, S.S.; Johansson, G. Characterization of Serine Protease Inhibitor from Solanum tuberosum Conjugated to Soluble Dextran and Particle Carriers. ACS Omega 2019, 4, 18456–18464.
- Durvanger, Z.; Boros, E.; Hegedus, R.; Dobo, J.; Kocsis, A.; Fodor, K.; Gal, P.; Mezo, G.; Pal, G.; Harmat, V.; et al. Studying the Structural Basis for Selectivity in Complexes of Peptide Inhibitors and Serine-Proteases of the Complement System. Acta. Crystallogr. A 2019, 75, E120.
- Shi, Y.H.; Shao, Y.N.; Lv, Z.M.; Li, C.H. Serpin-type serine protease inhibitor mediates coelomocyte apoptosis in Apostichopus japonicus. Fish Shellfish Immunol. 2020, 104, 410–418.
- Wei, X.M.; Xu, J.; Yang, J.M.; Liu, X.Q.; Zhang, R.R.; Wang, W.J.; Yang, J.L. Involvement of a Serpin serine protease inhibitor (OoSerpin) from mollusc Octopus ocellatus in antibacterial response. Fish Shellfish Immunol. 2015, 42, 79–87.
- Gettins, P.G.W. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4803.
- Hellinger, R.; Gruber, C.W. Peptide-based protease inhibitors from plants. Drug Discov. Today 2019, 24, 1877–1889.
- Huntington, J.A. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011, 9, 26–34.
- Turk, B.; Turk, D.; Turk, V. Protease signalling: The cutting edge. EMBO J. 2012, 31, 1630–1643.
- Bendre, A.D.; Ramasamy, S.; Suresh, C.G. Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int. J. Biol. Macromol. 2018, 113, 933–943.
- Blisnick, A.A.; Foulon, T.; Bonnet, S.I. Serine Protease Inhibitors in Ticks: An Overview of Their Role in Tick Biology and Tick-Borne Pathogen Transmission. Front. Cell. Infect. Microbiol. 2017, 7, 199.
- Gomes, M.T.; Oliva, M.L.; Lopes, M.T.; Salas, C.E. Plant proteinases and inhibitors: An overview of biological function and pharmacological activity. Curr. Protein Pept. Sci. 2011, 12, 417–436.
- Yang, X.; van der Donk, W.A. Ribosomally synthesized and post-translationally modified peptide natural products: New insights into the role of leader and core peptides during biosynthesis. Chemistry 2013, 19, 7662–7677.
- Armstrong, W.B.; Taylor, T.H.; Kennedy, A.R.; Melrose, R.J.; Messadi, D.V.; Gu, M.; Le, A.D.; Perloff, M.; Civantos, F.; Goodwin, W.J.; et al. Bowman birk inhibitor concentrate and oral leukoplakia: A randomized phase IIb trial. Cancer Prev. Res. 2013, 6, 410–418.
- Dai, H.; Ciric, B.; Zhang, G.X.; Rostami, A. Bowman-Birk Inhibitor attenuates experimental autoimmune encephalomyelitis by delaying infiltration of inflammatory cells into the CNS. Immunol. Res. 2011, 51, 145–152.
- Safavi, F.; Rostami, A. Role of serine proteases in inflammation: Bowman-Birk protease inhibitor (BBI) as a potential therapy for autoimmune diseases. Exp. Mol. Pathol. 2012, 93, 428–433.
- Palavalli, M.H.; Natarajan, S.S.; Wang, T.T.; Krishnan, H.B. Imbibition of soybean seeds in warm water results in the release of copious amounts of Bowman-Birk protease inhibitor, a putative anticarcinogenic agent. J. Agric. Food Chem. 2012, 60, 3135–3143.
- Zajc, I.; Sever, N.; Bervar, A.; Lah, T.T. Expression of cysteine peptidase cathepsin L and its inhibitors stefins A and B in relation to tumorigenicity of breast cancer cell lines. Cancer Lett. 2002, 187, 185–190.
- Martins, L.A.; Kotal, J.; Bensaoud, C.; Chmelar, J.; Kotsyfakis, M. Small protease inhibitors in tick saliva and salivary glands and their role in tick-host-pathogen interactions. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140336.
- Srikanth, S.; Chen, Z. Plant Protease Inhibitors in Therapeutics-Focus on Cancer Therapy. Front. Pharmacol. 2016, 7, 470.
- Filler, G.; Bokenkamp, A.; Hofmann, W.; Le Bricon, T.; Martinez-Bru, C.; Grubb, A. Cystatin C as a marker of GFR-history, indications, and future research. Clin. Biochem. 2005, 38, 1–8.
- Priyadarshini, M.; Khan, R.H.; Bano, B. Physicochemical properties of thiol proteinase inhibitor isolated from goat pancreas. Biopolymers 2010, 93, 708–717.
- Benchabane, M.; Schluter, U.; Vorster, J.; Goulet, M.C.; Michaud, D. Plant cystatins. Biochimie 2010, 92, 1657–1666.
- Vorster, B.J.; Goulet, M.C.; Michaud, D. Plant cystatins and insect cysteine proteases: Weapons in a molecular arms race. S. Afr. J. Bot. 2012, 79, 221–222.
- Farady, C.J.; Craik, C.S. Mechanisms of macromolecular protease inhibitors. Chembiochem A Eur. J. Chem. Biol. 2010, 11, 2341–2346.
- Gomis-Ruth, F.X.; Maskos, K.; Betz, M.; Bergner, A.; Huber, R.; Suzuki, K.; Yoshida, N.; Nagase, H.; Brew, K.; Bourenkov, G.P.; et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature 1997, 389, 77–81.
- Bateman, K.S.; James, M.N. Plant protein proteinase inhibitors: Structure and mechanism of inhibition. Curr. Protein Pept. Sci. 2011, 12, 340–347.
- Turra, D.; Lorito, M. Potato type I and II proteinase inhibitors: Modulating plant physiology and host resistance. Curr. Protein Pept. Sci. 2011, 12, 374–385.
- Li, B.J.; Gadahi, J.A.; Gao, W.X.; Zhang, Z.C.; Ehsan, M.; Xu, L.X.; Song, X.K.; Li, X.R.; Yan, R.F. Characterization of a novel aspartyl protease inhibitor from Haemonchus contortus. Parasites Vectors 2017, 10.
- Mondal, M.; Radeva, N.; Koster, H.; Park, A.; Potamitis, C.; Zervou, M.; Klebe, G.; Hirsch, A.K.H. Structure-Based Design of Inhibitors of the Aspartic Protease Endothiapepsin by Exploiting Dynamic Combinatorial Chemistry. Angew. Chem. Int. Edit. 2014, 53, 3259–3263.
- Motwani, H.V.; De Rosa, M.; Odell, L.R.; Hallberg, A.; Larhed, M. Aspartic protease inhibitors containing tertiary alcohol transition-state mimics. Eur. J. Med. Chem. 2015, 90, 462–490.
- Jiang, L.G.; Andersen, L.M.; Andreasen, P.A.; Chen, L.Q.; Huang, M.D. Insights into the serine protease mechanism based on structural observations of the conversion of a peptidyl serine protease inhibitor to a substrate. BBA Gen. Subj. 2016, 1860, 599–606.
- Zuchowski, J.; Grzywnowicz, K. Partial purification of proteinase K inhibitors from liquid-cultured mycelia of the white rot basidiomycete Trametes versicolor. Curr. Microbiol. 2006, 53, 259–264.
- Laskowski, M.; Qasim, M.A. What can the structures of enzyme-inhibitor complexes tell us about the structures of enzyme substrate complexes? BBA Protein Struct. M 2000, 1477, 324–337.
- Grosse-Holz, F.M.; van der Hoorn, R.A.L. Juggling jobs: Roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 2016, 210, 794–807.
- Clemente, M.; Corigliano, M.G.; Pariani, S.A.; Sanchez-Lopez, E.F.; Sander, V.A.; Ramos-Duarte, V.A. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345.
- Joshi, R.S.; Mishra, M.; Suresh, C.G.; Gupta, V.S.; Giri, A.P. Complementation of intramolecular interactions for structural-functional stability of plant serine proteinase inhibitors. BBA Gen. Subj. 2013, 1830, 5087–5094.
- Stoops, J.K.; Schroeter, J.P.; Kolodziej, S.J.; Strickland, D.K. Structure-Function-Relationships of Human Alpha(2)-Macroglobulin-3-Dimensional Structures of Native Alpha(2)-Macroglobulin and Its Methylamine and Chymotrypsin Derivatives. Biol. Alpha2 Macroglobulin Recept. Relat. Proteins 1994, 737, 212–228.
- Antao, C.M.; Malcata, F.X. Plant serine proteases: Biochemical, physiological and molecular features. Plant Physiol. Bioch. 2005, 43, 637–650.




