
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | sohail imran | + 1049 word(s) | 1049 | 2021-02-09 04:12:35 | | | |
| 2 | Catherine Yang | Meta information modification | 1049 | 2021-02-09 10:24:52 | | |
Video Upload Options
The bile salt export pump (BSEP/ABCB11) is responsible for the transport of bile salts from hepatocytes into bile canaliculi. Malfunction of this transporter results in progressive familial intrahepatic cholestasis type 2 (PFIC2), benign recurrent intrahepatic cholestasis type 2 (BRIC2) and intrahepatic cholestasis of pregnancy (ICP).
1. Introduction
The ATP-binding cassette (ABC) proteins constitute one of the largest families of membrane proteins. They are universally present in all kingdoms of life. In humans, 48 functional genes encode for ABC proteins, which on the basis of structural and sequence similarity are categorized into seven subfamilies, designated as ABCA through G [1]. Most of these proteins transport substrates across cellular membranes. A functional ABC transporter comprises at least four domains: two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs), as shown in Figure 1. In the ABCB subfamily, each of the TMDs consists of six membrane-spanning helices. Five of these extend into the cytoplasm to form an expansive intracellular domain. The two TMDs are responsible for substrate binding and translocation. The NBDs form two composite nucleotide-binding sites (NBSs) at their interface, which bind and hydrolyze ATP, and thereby provide the energy for substrate transport. These NBSs are formed by the Walker A and Walker B motifs, as well as the A-, Q- and H-loops of one NBD and the signature motif and D-loop of the other NBD [2][3].
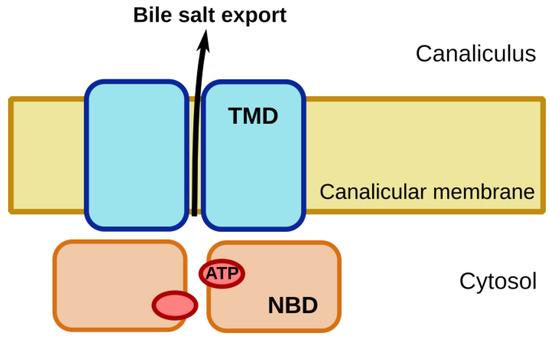
Figure 1. Schematic of the four-domain architecture of an ATP-binding cassette (ABC) transporter, as illustrated for the bile salt export pump BSEP. Hydrolysis of ATP by the nucleotide binding domains (NBD) energizes transport of bile salts from the hepatocyte cytoplasm to the bile canaliculus. Transmembrane domains (TMD) form the path for bile salt transport across the canalicular membrane.
The ABCB subfamily is one of the most diverse groups of ABC proteins, as it includes dimeric half transporters, monomers of which are each composed of one TMD and one NBD, but also full-length transporters, in which all four domains are fused into a single polypeptide chain. The former group comprises the homodimeric transporters ABCB6, ABCB7, ABCB8, ABCB9 and ABCB10, and the heterodimeric transporter ABCB2/ABCB3. The four full-length transporters of the ABCB subfamily are ABCB1, ABCB4, ABCB5 and ABCB11. The heterodimeric ABCB2/ABCB3 and full-length ABCB11 differ from the other members as they contain only one canonical NBS rather than two [4].
2. BSEP/ABCB11: Physiological Role
The bile salt export pump (BSEP) is expressed in hepatocytes. While high levels of BSEP mRNA were detected in testes and lower levels were reported in other extrahepatic tissues, including trachea, prostate, lungs, thymus, kidney and colon [5,6], plasma membrane expression of functional protein was found in liver cells only [5][6][7][8]. Adjacent hepatocytes form tight junctions to enclose functional structures called bile canaliculi, to which BSEP is targeted. Bile salts undergo an enterohepatic circulation, which depends on active transport systems in the liver and intestine. In the course of this process, newly synthesized and recycled bile salts are secreted from hepatocytes into bile canaliculi by BSEP and via bile ducts reach the duodenum. In the ileum, these bile salts are reabsorbed by the apical sodium-dependent bile salt transporter in intestinal epithelial cells (ASBT/SLC10A2). From the intestine, bile salts return to the liver via the superior mesenteric and portal veins, which carry the blood that feeds liver sinusoids. Uptake into hepatocytes is mediated by the sodium taurocholate co-transporting polypeptide (NTCP/SLC10A1) and organic anion transporters (OATPs). Bile salt transport by BSEP constitutes the rate-limiting step in bile formation and provides the major driving force for enterohepatic circulation [9].
The bile salt pool is recycled from the intestine to the liver six to eight times a day [10], resulting in daily bile salt excretion of about 20–40 g [11]. Impairment of BSEP results in the failure to maintain physiological bile flow, resulting in a clinical condition called intrahepatic cholestasis. BSEP has narrow specificity for its substrate bile salts, but the drugs pravastatin, vinblastine and fexofenadine are reported to be non-physiological substrates [12][13][14].
3. Transcriptional Regulation
BSEP expression is highly regulated by transcriptional mechanisms, and a wide inter-individual variability has been described at the mRNA and protein levels [15].
Expression of BSEP is regulated by a major ligand-activated transcription factor, farnesoid X receptor (FXR, NR1H4), which forms a signaling-competent nuclear receptor heterodimer with the retinoid X receptor (RXR) (Figure 2). Bile acids, such as chenodeoxycholic acid (CDCA), deoxycholic acid (DCA) and cholic acid (CA), are endogenous ligands of FXR with varying potential for activation [16][17][18][19]. Upon ligand binding, the FXR/RXR heterodimer binds to an FXR response element (FXRE) in the promoter region of BSEP, thereby inducing the expression of the transporter [20]. Additionally, components of the activating signal cointegrator-2-containing complex (ASCOM) interact with FXR to enhance BSEP expression. Ananthanarayanan and co-workers [21] showed that the recruitment of ASCOM to the BSEP promoter was disrupted in cholestasis, which was induced by common bile duct ligation. Furthermore, co-activator-associated arginine methyltransferase 1 (CARM1) also regulates FXR/RXR-dependent BSEP transcription [22]. Similarly, steroid receptor co-activator 2 (SRC2) knockout mice showed reduced expression of BSEP [23], indicating its involvement in transcriptional regulation of the transporter.
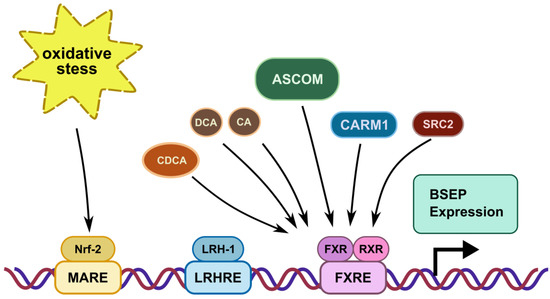
Figure 2. Transcriptional regulation of BSEP via recognition sequences in the promoter region of the BSEP gene: MARE: Maf recognition element; LRHRE: liver receptor homolog-1 responsive element; FXRE: farnesoid X receptor responsive element. The farnesoid receptor FXR binds bile salts after heterodimerization with the retinoid X receptor. The ligand with the highest affinity for FXR is chenodeoxycholic acid (CDCA): however deoxycholic (DCA) and cholic acid (CA) also increase BSEP expression. The co-activators ‘activating signal cointegrator-2-containing complex’ (ASCOM), ‘co-activator-associated arginine methyltransferase 1’ (CARM1) and ‘steroid receptor co-activator SRC2’ (SRC2) increase BSEP expression via FXR. Liver receptor homolog 1 (LHR-1) and nuclear factor erythroid 2-related factor (Nrf-2), a sensor for oxidative stress, also increase BSEP expression.
Hepatocyte-specific liver receptor homolog-1 (LRH-1, NR5A2) is another transcription factor involved in modulation of BSEP expression. LRH-1 plays a supporting role for FXR [24]. The absence of LRH-1 is associated with reduced BSEP expression and an altered BA composition, with disappearance of CA and taurocholic acid (TCA) [25]. BSEP promoter activity is also stimulated by nuclear factor erythroid 2-related factor 2 (Nrf2), a positive transcriptional regulator, which acts as a sensor for oxidative stress. Nrf2 regulates the expression of BSEP, but also that of a number of hepatic phase I and II enzymes and other hepatic efflux transporters such as MRP3 (ABCC3) and MRP4 (ABCC4) [26].
References
- Dean, M.; Rzhetsky, A.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166.
- Schmitt, L. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2002, 12, 754–760.
- Shintre, C.A.; Pike, A.C.W.; Li, Q.; Kim, J.-I.; Barr, A.J.; Goubin, S.; Shrestha, L.; Yang, J.; Berridge, G.; Ross, J.; et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. USA 2013, 110, 9710–9715.
- Procko, E.; O’Mara, M.L.; Bennett, W.F.D.; Tieleman, D.P.; Gaudet, R. The mechanism of ABC transporters: General lessons from structural and functional studies of an antigenic peptide transporter. FASEB J. 2009, 23, 1287–1302.
- Langmann, T.; Mauerer, R.; Zahn, A.; Moehle, C.; Probst, M.; Stremmel, W.; Schmitz, G. Real-Time Reverse Transcription-PCR Expression Profiling of the Complete Human ATP-Binding Cassette Transporter Superfamily in Various Tissues. Clin. Chem. 2003, 49, 230–238.
- Stieger, B. The Role of the Sodium-Taurocholate Cotransporting Polypeptide (NTCP) and of the Bile Salt Export Pump (BSEP) in Physiology and Pathophysiology of Bile Formation. In Drug Transporters; Springer: Berlin/Heidelberg, Germany, 2011; pp. 205–259.
- Lagana, S.M.; Salomao, M.; Remotti, H.E.; Knisely, A.S.; Moreira, R.K. Bile salt export pump: A sensitive and specific immunohistochemical marker of hepatocellular carcinoma. Histopathology 2015, 66, 598–602.
- Hilgendorf, C.; Ahlin, G.; Seithel, A.; Artursson, P.; Ungell, A.-L.; Karlsson, J. Expression of Thirty-six Drug Transporter Genes in Human Intestine, Liver, Kidney, and Organotypic Cell Lines. Drug Metab. Dispos. 2007, 35, 1333–1340.
- Stieger, B.; Beuers, U. The Canalicular Bile Salt Export Pump BSEP (ABCB11) as a Potential Therapeutic Target. Curr. Drug Targets 2011, 12, 661–670.
- Trauner, M.; Fuchs, C.D.; Halilbasic, E.; Paumgartner, G. New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology 2017, 65, 1393–1404.
- Trauner, M.; Boyer, J.L. Bile Salt Transporters: Molecular Characterization, Function, and Regulation. Physiol. Rev. 2003, 83, 633–671.
- Lecureur, V.; Sun, D.; Hargrove, P.; Schuetz, E.G.; Kim, R.B.; Lan, L.B.; Schuetz, J.D. Cloning and expression of murine sister of P-glycoprotein reveals a more discriminating transporter than MDR1/P-glycoprotein. Mol. Pharmacol. 2000, 57, 24–35.
- Hirano, M.; Maeda, K.; Hayashi, H.; Kusuhara, H.; Sugiyama, Y. Bile Salt Export Pump (BSEP/ABCB11) Can Transport a Nonbile Acid Substrate, Pravastatin. J. Pharmacol. Exp. Ther. 2005, 314, 876–882.
- Matsushima, S.; Maeda, K.; Hayashi, H.; Debori, Y.; Schinkel, A.H.; Schuetz, J.D.; Kusuhara, H.; Sugiyama, Y. Involvement of Multiple Efflux Transporters in Hepatic Disposition of Fexofenadine. Mol. Pharmacol. 2008, 73, 1474–1483.
- Ho, R.H.; Leake, B.F.; Kilkenny, D.M.; Meyer zu Schwabedissen, H.E.; Glaeser, H.; Kroetz, D.L.; Kim, R.B. Polymorphic variants in the human bile salt export pump (BSEP; ABCB11): Functional characterization and interindividual variability. Pharmacogenet. Genom. 2010, 20, 45–57.
- Makishima, M. Identification of a Nuclear Receptor for Bile Acids. Science 1999, 284, 1362–1365.
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous Bile Acids Are Ligands for the Nuclear Receptor FXR/BAR. Mol. Cell 1999, 3, 543–553.
- Parks, D.J. Bile Acids: Natural Ligands for an Orphan Nuclear Receptor. Science 1999, 284, 1365–1368.
- Baghdasaryan, A.; Chiba, P.; Trauner, M. Clinical application of transcriptional activators of bile salt transporters. Mol. Aspects Med. 2014, 37, 57–76.
- Plass, J.R.M.; Mol, O.; Heegsma, J.; Geuken, M.; Faber, K.N.; Jansen, P.L.M.; Müller, M. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology 2002, 35, 589–596.
- Ananthanarayanan, M.; Li, Y.; Surapureddi, S.; Balasubramaniyan, N.; Ahn, J.; Goldstein, J.A.; Suchy, F.J. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am. J. Physiol. Liver Physiol. 2011, 300, G771–G781.
- Ananthanarayanan, M.; Li, S.; Balasubramaniyan, N.; Suchy, F.J.; Walsh, M.J. Ligand-dependent Activation of the Farnesoid X-receptor Directs Arginine Methylation of Histone H3 by CARM1. J. Biol. Chem. 2004, 279, 54348–54357.
- Chopra, A.R.; Kommagani, R.; Saha, P.; Louet, J.-F.; Salazar, C.; Song, J.; Jeong, J.; Finegold, M.; Viollet, B.; DeMayo, F.; et al. Cellular Energy Depletion Resets Whole-Body Energy by Promoting Coactivator-Mediated Dietary Fuel Absorption. Cell Metab. 2011, 13, 35–43.
- Song, X.; Kaimal, R.; Yan, B.; Deng, R. Liver receptor homolog 1 transcriptionally regulates human bile salt export pump expression. J. Lipid Res. 2008, 49, 973–984.
- Mataki, C.; Magnier, B.C.; Houten, S.M.; Annicotte, J.-S.; Argmann, C.; Thomas, C.; Overmars, H.; Kulik, W.; Metzger, D.; Auwerx, J.; et al. Compromised Intestinal Lipid Absorption in Mice with a Liver-Specific Deficiency of Liver Receptor Homolog 1. Mol. Cell. Biol. 2007, 27, 8330–8339.
- Weerachayaphorn, J.; Cai, S.-Y.; Soroka, C.J.; Boyer, J.L. Nuclear factor erythroid 2-related factor 2 is a positive regulator of human bile salt export pump expression. Hepatology 2009, 50, 1588–1596.




