| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nikola Puvača | + 7241 word(s) | 7241 | 2021-02-08 09:58:42 | | | |
| 2 | Rita Xu | + 3329 word(s) | 3329 | 2021-02-08 10:19:05 | | |
Video Upload Options
Public healthcare systems all over the world are faced with a great challenge in this respect. Obviously, there are many bacteria that can cause infections in humans and animals alike, but somehow it seems that the greatest threat nowadays comes from the Enterobacteriaceae members, especially Escherichia coli.
1. Introduction
Scientists all over the world have studied Escherichia coli and it appears to be the most thoroughly investigated and best understood of all model microorganisms [1][2][3][4]. We already know that it is one of the first bacteria that colonizes the human gut immediately after birth [5][6][7]. On the other hand, E. coli is often the main culprit of infections in the gastrointestinal tract [8], as well as other parts of human and animal organisms [9][10]. In more precise terms, E. coli typically causes urinary infections [11][12], but it can also lead to many other serious infections and conditions, such as: appendicitis [13], pneumonia [14], meningitis [15], endocarditis [16], gastrointestinal infections [17], etc. Research findings have shown us that E. coli can cause infections in all age groups and those infections can be acquired in the general population, i.e., community-acquired, as well as related to healthcare institutions [18][19][20].
After Alexander Fleming had discovered penicillin in 1928, the whole course of medicine changed [21][22]. The revolutionary discovery of antibiotics made it possible for doctors to treat extremely severe cases of infectious diseases, which had previously been a very common cause of death [23][24]. That completely changed after antibiotics had been introduced and soon penicillin became the most widely used antibiotic in the world, saving millions of lives [25][26][27].
Unfortunately, only several years after doctors started using it in hospitals, the first cases of penicillin resistance by Staphylococcus aureus were identified [28]. Obviously, bacteria have managed to develop a system that can protect them and make them resistant to antibiotics [29]. Sadly, the situation with bacteria evolving resistance is getting worse day by day and we have literally come to a point when we can speak of the antimicrobial resistance presenting a worldwide problem [30][31][32][33][34][35][36].
When we speak about E. coli, the fact that it has been put on the World Health Organization’s (WHO) list that contains 12 families of bacteria that present the biggest danger to human health [37][38]. Ever since the first reported cases, E. coli’s resistance to antibiotic treatment has been continuously growing [39][40][41][42].
Scientific literature offers an abundance of research studies into the nature and behavior of E. coli [43][44][45][46]. The results point to several extremely interesting facts. This bacterium undoubtedly has considerable influence on human and animal lives [47][48], for the simple reason that it lives inside the gut and can very easily spread from fecal matter to the mouth [49][50]. Being the commensal bacteria of human and animal gut, it happens to be in close contact with numerous other bacteria [51]. However, perhaps the most fascinating thing about E. coli is its ability to pass on its genetic-resistant traits to microorganisms who share the same living environment, as well as to acquire resistance genes from them [52][53][54].
According to Poirel et al. [52] E. coli present a bacterium with a special place in the microbiological world since it can cause severe infections in humans and animals, and on the other hand represents a significant part of the autochthonous microbiota of the various hosts. The main apprehension is a transmission of virulent and resistant E. coli among animals and humans through various pathways. E. coli is a most important reservoir of resistance genes that may be accountable for treatment failures in both human and veterinary medicine [52]. An increasing number of resistance genes has been identified in E. coli isolates in the past 10 years, and many of these resistance genes were acquired by horizontal gene transfer. In the enterobacterial gene pool, E. coli acts as a donor and as a recipient of resistance genes and thereby can acquire resistance genes from other bacteria but can also pass on its resistance genes to other bacteria. Antimicrobial resistance in E. coli is considered one of the foremost disputes in both humans and animals at a global scale and needs to be considered as a real public health concern.
Barrios-Villa et al. [55] have observed increased evidence demonstrating the association between Crohn’s Disease (CD), a type of Inflammatory Bowel Disease (IBD), and non-diarrheagenic Adherent/Invasive E. coli (AIEC) isolates. Genomes of five AIEC strains isolated from individuals without IBD were sequenced and compared with AIEC prototype strains (LF82 and NRG857c), and with extra-intestinal uropathogenic strain (UPEC CFT073). Non-IBD-AIEC strains showed an Average Nucleotide Identity up to 98% compared with control strains. Blast identities of the five non-IBD-AIEC strains were higher when compared to AIEC and UPEC reference strains than with another E. coli pathotypes, suggesting a relationship between them [55]. In the same study, Barrios-Villa et al. [55], an incomplete Type VI secretion system was found in non-IBD-AIEC strains; however, the Type II secretion system was complete. Several groups of genes reported in AIEC strains were searched in the five non-IBD-AIEC strains, and the presence of fimA, fliC, fuhD, chuA, irp2, and cvaC were confirmed. Other virulence factors were detected in non-IBD-AIEC strains, which were absent in AIEC reference strains, including EhaG, non-fimbrial adhesin 1, PapG, F17D-G, YehA/D, FeuC, IucD, CbtA, VgrG-1, Cnf1, and HlyE. Based on the differences in virulence determinants and single-nucleotide polymorphisms (SNPs), it is plausible to suggest that non-IBD AIEC strains belong to a different pathotype.
Meanwhile, genomic analysis of E. coli strains isolated from diseased chicken in the Czech Republic [56] showed that multiresistant phenotype was detected in most of the sequenced strains with the predominant resistance to β-lactams and quinolones being associated with TEM-type beta-lactamase genes and chromosomal gyrA mutations. The phylogenetic analysis proved a huge variety of isolates that were derived from all groups. Clusters of closely related isolates within ST23 and ST429 indicated a possible local spread of these clones. Moreover, the ST429 cluster carried blaCMY-2,− 59 genes for AmpC β-lactamase and isolates of both clusters were well-equipped with virulence-associated genes, with significant variations in allocation of specific virulence-associated genes among phylogenetically distant lineages. Zoonotic APEC STs were also identified, such as ST117, ST354, and ST95, showing numerous molecular elements typical for human ExPEC [56].
As already stated, antibiotic resistance found in microorganisms presents a big challenge for medical practice in the whole world [57][58][59][60][61]. This is to a great extent the consequence of wrong or uncritical consumption of antibiotics.
In a study by Abdelhalim et al. [62], from 17 Crohn’s disease patients and 14 healthy controls E. coli strains were isolated, 59% and 50% of them were identified as AIEC strains. It was discovered that chuA and ratA genes were the most significant genetic markers associated with AIEC compared to non-AIEC strains isolated from Crohn’s disease patients and healthy controls p = 0.0119, 0.0094, respectively. Most E. coli strains obtained from Crohn’s disease patients showed antibiotic resistance (71%) compared to healthy controls (29%) against at least one antibiotic. Investigation have demonstrated significant differences between AIEC strains and non-AIEC strains in terms of the prevalence of chuA and ratA virulence genes and the antibiotic resistance profiles. Furthermore, AIEC strains isolated from Crohn’s disease patients were found to be more resistant to β-lactam and aminoglycoside antibiotics than AIEC strains isolated from healthy controls [62].
E. coli strains isolated from animals in Tunisia [63] revealed occurrence of plasmid-mediated quinolone resistance between themselves. With 51 nalidixic acid-resistant isolates, 9 PMQR genes were harbored (5 co-harbored qnrS1 and qnrB1, 3 harbored qnrS1 and 1 harbored qnrB1). Two types of mutation in the QRDR of GyrA were observed: S83L and D87N. For the QRDR of ParC, the substitution S80I was observed as well, while A class 1 integron was found in isolates, respectively. The tetA or tetB gene was observed and both were co-harbored by two isolates. The sul1, sul2, and sul3 genes were discovered, respectively. According to the presence of specific virulence genes, the nine strains were classified as UPEC, EAEC, and EPEC [63]. All mentioned highlight the plausible role of the avian industry as a reservoir of human pathogenic E. coli strains.
Yu et al. [64] have investigated the prevalence and antimicrobial-resistance phenotypes and genotypes of E. coli isolated from raw milk samples from mastitis cases in four regions of China. A total of 83 strains of E. coli were isolated and identified, but without any significant differences in the number of E. coli isolates detected among the two sampling seasons in the same regions. Nevertheless, a significant difference in E. coli prevalence was found among the four different regions. The isolates were most frequently resistant to penicillin (100%), acetylspiramycin (100%), lincomycin (98.8%), oxacillin (98.8%), and sulphamethoxazole (53%). All the E. coli strains were multiresistant to three antimicrobial classes, and the most frequent multidrug-resistance patterns for the isolates were resistant to three or four classes of drugs simultaneously [64].
In Egypt, Farhat et al. [65] have investigated the antimicrobial resistance patterns, the distribution of phylogenetic groups, and the prevalence and characteristics of integron-bearing E. coli isolates from outpatients with community-acquired urinary tract infections. A total of 134 human urine samples were positive for E. coli, from which a total of 80 samples were selected for further analyses. Most of the isolates (62.5%) proved multidrug resistance profiles. Group B2 was the most predominant phylogenetic group (52.5%), followed by group F (21.25%), Clade I or II (12.5%), and finally isolates of unknown phylogroup (13.75%). Of the 80 selected isolates, 7 of them carried class 1 integrons, which contained 3 different types of integrated gene cassettes, conferring resistance to streptomycin, trimethoprim, and some open reading frames of unknown function [65].
Low hygiene levels, lack of clean water, or poor sanitary conditions can create perfect conditions for the development and transmission of infections [66]. In addition to that, Farhani et al. [67] have total of 80 E. coli isolates, separated into 51 different genotypes. Using the Multi Locus VNTR Analysis (MLVA) profiles, a minimum spanning tree (MST) algorithm showed two clonal complexes with 71 isolates and only 9 isolates were stayed out of clonal complexes in the form of a singleton. High genotypic diversity was seen among E. coli strains isolated from hospital wastewaters; however, many isolates showed a close genetic relationship. Authors have concluded that MLVA as a rapid, inexpensive, and useful tool could be used for analysis of the phylogenetic relationships between E. coli strains [67].
Extended-spectrum beta-lactamases (ESBLs) are specific enzymes, which show resistance to almost all beta-lactam antibiotics [68], including penicillin [69], cephalosporin [70], etc. [71]. Cases of infections in which ESBLs are produced usually have quite an unpredictable course. E. coli is an example of a multidrug-resistant and ESBL-producing bacterium that can be the source of extremely severe infections [72][73][74]. As has previously been stated, some strains of E. coli can also cause very serious medical conditions connected with urinary and gastrointestinal tract and central nervous system [75]. On the other hand, the side effects of a prolonged usage of antibiotics include the occurrence of antibiotic resistance [76][77][78]. Today we have evidence that people can get antibiotic-resistant E. coli directly or indirectly from the environment [79][80]. Therefore, it is very important that we first evaluate the existence of drug-resistant E. coli in our surroundings and based on such findings try to outline the human and veterinary healthcare guidelines [81][82][83][84][85][86].
2. Usage of Antibiotics in Different Countries of EU Region and Spread of E. coli Resistance to Antibiotics
It is absolutely clear to us today that the antibiotic resistance of E. coli and some other bacteria involves a combination of different factors [87][88]. Research results indicate that E. coli exhibits the strongest resistance to the longest used and most commonly prescribed antibiotics [89][90][91]. This is exactly the case with sulfonamides, which were first used in humans around 1930s [92]. Some twenty years later, the first resistant strains of E. coli appeared and with time this resistance only grew stronger. It has also been found that low-income [93] and mid-income countries (Table 1) are regions with the highest antibiotic-resistance rates and it is precisely in these regions that we see the highest consumption of antibiotics [94]. On the other hand, high-income nations show a lower rate of antibiotic resistance, resulting from lower usage of antibiotics. In some high-income countries the consumption is high, for example in Belgium, France, and Italy. This is even more complex when comparing to low-income countries where on one hand the consumption may be high but the availability of many of the more advanced antimicrobials is limited [95].
Table 1. The consumption of total antibiotics in Defined Daily Doses, in DDD per 1000 inhabitants per day in countries of European region based on WHO database [93].
|
Country |
DDD/1000 Inhabitants Per Day |
Country |
DDD/1000 Inhabitants Per Day |
|
Albania |
16.41 |
Kosovo |
20.18 |
|
Armenia |
10.31 |
Kyrgyzstan |
17.94 |
|
Austria |
12.17 |
Latvia |
13.30 |
|
Azerbaijan |
7.66 |
Lithuania |
15.83 |
|
Belarus |
17.48 |
Luxemburg |
22.31 |
|
Belgium |
25.57 |
Malta |
21.88 |
|
Bosnia and Herzegovina |
17.85 |
Montenegro |
29.33 |
|
Bulgaria |
20.25 |
Netherlands |
9.78 |
|
Croatia |
20.28 |
Norway |
16.97 |
|
Cyprus |
27.14 |
Poland |
24.30 |
|
Czech Republic |
17.18 |
Portugal |
17.72 |
|
Denmark |
17.84 |
North Macedonia |
13.42 |
|
Estonia |
12.13 |
Romania |
28.50 |
|
Finland |
18.52 |
Russia |
14.82 |
|
France |
25.92 |
Serbia |
31.57 |
|
Georgia |
24.44 |
Slovakia |
24.34 |
|
Germany |
11.49 |
Slovenia |
13.48 |
|
Greece |
33.85 |
Spain |
17.96 |
|
Hungary |
16.31 |
Sweden |
13.73 |
|
Iceland |
17.87 |
Tajikistan |
21.95 |
|
Ireland |
23.27 |
Turkey |
38.18 |
|
Italy |
26.62 |
United Kingdom |
20.47 |
|
Kazakhstan |
17.89 |
Uzbekistan |
8.56 |
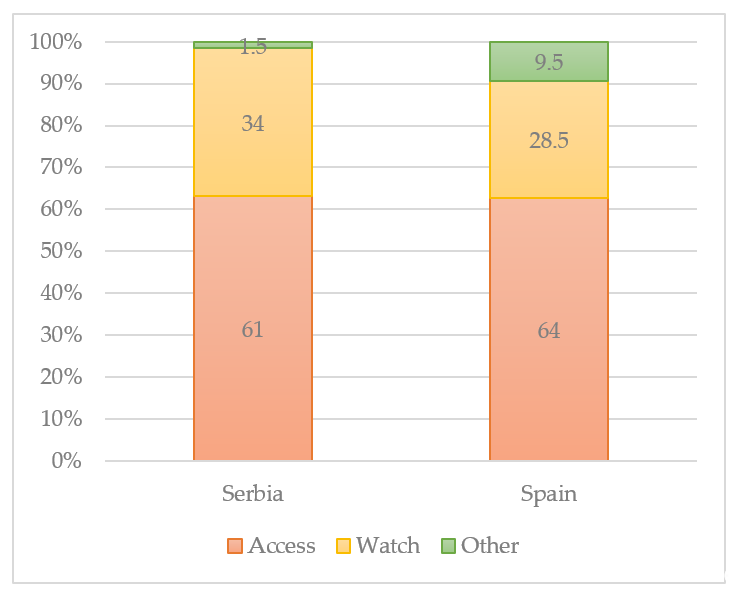
In the 2017 revision of the WHO Model List of Essential Medicines, antibiotics in the list were grouped into three AWaRe categories: Access, Watch, and Reserve. According to the WHO AWaRe categories [96], the classification showed that the Access group antibiotics accounted for more than 50% of total consumption both in Serbia and Spain [93]. The size of the population (in thousands) living in the European Region in 2015 was 912,984, respectively. Of the 53 Member States of the region, none is a low-income country, 20 are middle-income countries, and 33 are high-income countries. The median proportional consumption of the Access group values ranged between 61% in Spain to 64% in Serbia. The median proportion of Watch group antibiotics related to total consumption values ranging from less than 34% in Serbia and 28.5% in Spain. Reserve group antibiotics were only rarely used. The most widely used Reserve group antibiotics were intravenous fosfomycin, followed by cefepime, colistin, linezolid, and daptomycin. The antibiotics assigned to the Other group varied from 1.5% in Serbia to 9.5% in Spain (Figure 1). Overall consumption of antibiotics in these 46 countries ranged from 7.66 to 38.18 DDD per 1000 inhabitants per day. The overall absolute weight (not adjusted by population size) varied from 2.18 ton (Iceland) to 1195.69 tons (Turkey) per year.
Figure 1. Proportional consumption of antibiotics by AWaRe categorization, % [93][96].
It is a widespread opinion among scientists that antibiotic resistance has developed as the result of human activity and commonly applied treatment with antibiotics [97]. On the other hand, studies of bacteria living inside human body and other environmental bacteria helped us discover many other resistance factors that did not develop over time as a reaction to antibiotics, but were probably part of bacteria genomes in the first place [98][99][100]. Scientists often refer to those characteristics as the intrinsic resistance of bacteria [101]. It presents a great advantage of that particular bacteria strain, as its main task is to inhibit or eliminate other bacteria that live in the same environment and compete for food [102][103][104]. Hence, intrinsic resistance is different from the extrinsic antibiotic resistance, which was triggered primarily by human action [105]. In times of constantly growing antibiotic resistance and in a situation when we seem not to have any readily available antibacterial agents, it is extremely important to thoroughly study the intrinsic resistance of bacteria. That could lead to the development of a new method of fight against bacterial resistance [106]. If we could manage somehow to inhibit the factors that intrinsic resistance is composed of, perhaps bacteria would then become highly sensitive to antibiotics again. E. coli and other gram-negative bacteria have two important characteristics, which are the foundations of their intrinsic resistance. Namely, they have a protective impermeable membrane and a large number of efflux pumps, which successfully remove all unwanted substances from inside the cell [107][108][109].
Antibiotic resistance is an ecosystem problem threatening the interrelated human–animal–environment health under the “One Health” framework. Resistant bacteria arising in one geographical area can spread via cross-reservoir transmission to other areas worldwide either by direct exposure or through the food chain and the environment. Drivers of antibiotic resistance are complex and multisectoral particularly in lower- and middle-income countries. These include inappropriate socio-ecological behaviors; poverty; overcrowding; lack of surveillance systems; food supply chain safety issues; highly contaminated waste effluents; and loose rules and regulations. Iskandar et al. [110] have investigated the drivers of antibiotic resistance from a “One Health” perspective. They have summarized the results from many researches that have been conducted over the years and shown that the market failures are the leading cause for the negative externality of antibiotic resistance that extends in scope from the individual to the global ecosystem. Iskandar et al. [110] highlighted that the problem will continue to prevail if governments do not prioritize the “One Health” approach and if individual’s accountability is still denied in a world struggling with profound socio-economic problems.
Dsani et al. [111] investigated the spread of E. coli isolates from raw meat in Greater Accra region in Ghana, to antibiotics resistance, respectively. Usually, raw meat can be contaminated with antibiotic resistant pathogens and consumption of meat contaminated with antibiotic resistant E. coli is associated with grave health care consequences. In their research, E. coli was detected in half of raw meat samples. Isolates were resistant to ampicillin (57%), tetracycline (45%), sulfamethoxazole-trimethoprim (21%), and cefuroxime (17%). Multidrug resistance (MDR) was identified in 22% of the isolates. The blaTEM gene was detected in 4% of E. coli isolates [111]. Dsani et al. [111] concluded that levels of microbial contamination of raw meat in their research were unacceptable and highlighted that meat handlers and consumers are at risk of foodborne infections from E. coli including ESBL producing E. coli, which is resistant to nearly all antibiotics in use.
According to Hassan et al. [112], a last resort antibiotic is colistin. Colistin is crucial for managing infections with carbapenem-resistant Enterobacteriaceae. The recent emergence of mobile-colistin-resistance (mcr) genes has jeopardized the efficiency of this antibiotic. Aquaculture is a foremost contributor to the evolution and dissemination of mcr. Nevertheless, data on mcr in aquaculture are narrow. In Lebanon, a country with developed antimicrobial stewardship the occurrence of mcr-1 was evaluated in fish. Mobile-colistin-resistance-1 was detected in 5 E. coli isolated from fish intestines. The isolates were classified as multidrug-resistant and their colistin minimum inhibitory concentration ranged between 16 and 32 μg/mL. Whole genome sequencing analysis showed that mcr-1 was carried on transmissible IncX4 plasmids and that the isolates harbored more than 14 antibiotic resistance genes. The isolates belonged to ST48 and ST101, which have been associated with mcr and can occur in humans and fish and help in spreading of antibiotic resistance of E. coli.
While, Montealegre et al. [113] have showed how high genomic diversity and heterogeneous origins of pathogenic and antibiotic-resistant E. coli in household settings represent a challenge to reducing transmission in low-income settings. Transmission of E. coli between hosts and with the environment is believed to happen more frequently in regions with poor sanitation. Montealegre et al. [113] performed whole-genome comparative analyses on 60 E. coli isolates from soils and fecal from cattle, chickens, and humans, in households in rural Bangladesh. Results suggest that in rural Bangladesh, a high level of E. coli in soil is possible led by contributions from multiple and diverse E. coli sources (human and animal) that share an accessory gene pool relatively unique to previously published E. coli genomes. Thus, interventions to reduce environmental pathogen or antimicrobial resistance transmission should adopt integrated “One Health” approaches that consider heterogeneous origins and high diversity to improve effectiveness and reduce prevalence and transmission [113].
It has been confirmed that wastewater treatment plant effluents are influenced by hospital wastewaters [114] in Germany. Alexander et al. [114] quantified the abundances of antibiotic resistance genes and facultative pathogenic bacteria as well as one mobile genetic element in genomic DNA via qPCR from 23 different wastewater treatment plant effluents in Germany. Total of 12 clinically relevant antibiotic resistance genes were categorized into frequently, intermediately, and rarely occurring genetic parameters of communal wastewaters. Taxonomic PCR quantifications of 5 facultative pathogenic bacteria targeting E. coli, P. aeruginosa, K. pneumoniae, A. baumannii, and enterococci were performed.
Since communal wastewater treatment plants are the direct link to the aquatic environment, wastewater treatment plants should be monitored according to their antibiotic resistance genes and facultative pathogenic bacteria abundances and discharges to decide about the need of advanced treatment options. Critical threshold volumes of hospital wastewaters should be defined to discuss the effect of a decentralized wastewater treatment, because they can serve as an excellent reservoir in spreading of E. coli resistance to antibiotic.
References
- Hill, R.A.; Hunt, J.; Sanders, E.; Tran, M.; Burk, G.A.; Mlsna, T.E.; Fitzkee, N.C. Effect of Biochar on Microbial Growth: A Metabolomics and Bacteriological Investigation in Coli. Environ. Sci. Technol. 2019, 53, 2635–2646, doi:10.1021/acs.est.8b05024.
- Lovley, D.R.; Holmes, D.E. Protein Nanowires: The Electrification of the Microbial World and Maybe Our Own. Bacteriol. 2020, 202, e00331-20, doi:10.1128/JB.00331-20.
- Ranganathan, S.; Smith, E.M.; Abel, J.D.F.; Barry, E.M. Research in a Time of Enteroids and Organoids: How the Human Gut Model Has Transformed the Study of Enteric Bacterial Pathogens. Gut Microbes. 2020, 12, 1795492, doi:10.1080/19490976.2020.1795389.
- Macklin, D.N.; Horst, T.A.A.; Choi, H.; Ruggero, N.A.; Carrera, J.; Mason, J.C.; Sun, G.; Agmon, E.; DeFelice, M.M.; Maayan, I.; et al. Simultaneous Cross-Evaluation of Heterogeneous E. Coli Datasets via Mechanistic Simulation. Science 2020, 369, eaav3751, doi:10.1126/science.aav3751.
- Micenková, L.; Bosák, J.; Smatana, S.; Novotný, A.; Budinská, E.; Šmajs, D. Administration of the Probiotic Escherichia Coli Strain A0 34/86 Resulted in a Stable Colonization of the Human Intestine During the First Year of Life. Antimicrob. Prot. 2020, 12, 343–350, doi:10.1007/s12602-019-09548-3.
- Bittinger, K.; Zhao, C.; Li, Y.; Ford, E.; Friedman, E.S.; Ni, J.; Kulkarni, C.V.; Cai, J.; Tian, Y.; Liu, Q.; et al. Bacterial Colonization Reprograms the Neonatal Gut Metabolome. Microbiol. 2020, 5, 838–847, doi:10.1038/s41564-020-0694-0.
- Secher, T.; Brehin, C.; Oswald, E. Early Settlers: Which Coli Strains Do You Not Want at Birth? Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G123–G129, doi:10.1152/ajpgi.00091.2016.
- Rossi, E.; Cimdins, A.; Lüthje, P.; Brauner, A.; Sjöling, Å.; Landini, P.; Römling, U. “It’s a Gut Feeling”—Escherichia Coli Biofilm Formation in the Gastrointestinal Tract Environment. Rev. Microbiol. 2018, 44, 1–30, doi:10.1080/1040841X.2017.1303660.
- Zhang, S.; Abbas, M.; Rehman, M.U.; Huang, Y.; Zhou, R.; Gong, S.; Yang, H.; Chen, S.; Wang, M.; Cheng, A. Dissemination of Antibiotic Resistance Genes (ARGs) via Integrons in Escherichia Coli: A Risk to Human Health. Pollut. 2020, 266, 115260, doi:10.1016/j.envpol.2020.115260.
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. Trop. Med. 2020, 2020, 4674235, doi:10.1155/2020/4674235. 4674235
- Isla, A.L.; Polo, J.M.; Isa, M.A.; Sala, R.B.; Sutil, R.S.; Quintas, J.J.; Moradillo, J.G.; Padilla, D.A.G.; Rojo, E.G.; Martínez, J.B.P.; et al. Urinary Infections in Patients with Catheters in the Upper Urinary Tract: Microbiological Study. Int. 2017, 98, 442–448, doi:10.1159/000467398.
- Rodrigues, W.; Miguel, C.; Nogueira, A.; Vieira, C.U.; Paulino, T.; Soares, S.; De Resende, E.; Chica, J.L.; Araújo, M.; Oliveira, C. Antibiotic Resistance of Bacteria Involved in Urinary Infections in Brazil: A Cross-Sectional and Retrospective Study. Ijerph 2016, 13, 918, doi:10.3390/ijerph13090918.
- Song, D.W.; Park, B.K.; Suh, S.W.; Lee, S.E.; Kim, J.W.; Park, J.-M.; Kim, H.R.; Lee, M.-K.; Choi, Y.S.; Kim, B.G.; et al. Bacterial Culture and Antibiotic Susceptibility in Patients with Acute Appendicitis. J. Colorectal Dis. 2018, 33, 441–447, doi:10.1007/s00384-018-2992-z.
- Park, J.; Kim, S.; Lim, H.; Liu, A.; Hu, S.; Lee, J.; Zhuo, H.; Hao, Q.; Matthay, M.A.; Lee, J.-W. Therapeutic Effects of Human Mesenchymal Stem Cell Microvesicles in an Ex Vivo Perfused Human Lung Injured with Severe Coli Pneumonia. Thorax 2019, 74, 43–50, doi:10.1136/thoraxjnl-2018-211576.
- Zhao, W.-D.; Liu, D.-X.; Wei, J.-Y.; Miao, Z.-W.; Zhang, K.; Su, Z.-K.; Zhang, X.-W.; Li, Q.; Fang, W.-G.; Qin, X.-X.; et al. Caspr1 Is a Host Receptor for Meningitis-Causing Escherichia Coli. Commun. 2018, 9, 2296, doi:10.1038/s41467-018-04637-3.
- Akuzawa, N.; Kurabayashi, M. Native Valve Endocarditis Due to Escherichia Coli Infection: A Case Report and Review of the Literature. BMC Cardiovasc. Disord. 2018, 18, 195, doi:10.1186/s12872-018-0929-7.
- Sarowska, J.; Koloch, B.F.; Kmiecik, A.J.; Madrzak, M.F.; Ksiazczyk, M.; Ploskonska, G.B.; Krol, I.C. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia Coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019, 11, 10, doi:10.1186/s13099-019-0290-0.
- Poolman, J.T.; Anderson, A.S. Escherichia Coli and Staphylococcus Aureus: Leading Bacterial Pathogens of Healthcare Associated Infections and Bacteremia in Older-Age Populations. Expert Rev. Vaccines 2018, 17, 607–618, doi:10.1080/14760584.2018.1488590.
- Kubone, P.Z.; Mlisana, K.P.; Govinden, U.; Abia, A.L.K.; Essack, S.Y. Antibiotic Susceptibility and Molecular Characterization of Uropathogenic Escherichia Coli Associated with Community-Acquired Urinary Tract Infections in Urban and Rural Settings in South Africa. TropicalMed 2020, 5, 176, doi:10.3390/tropicalmed5040176.
- Djordjevic, Z.; Folic, M.; Jankovic, S. Community-Acquired Urinary Tract Infections: Causative Agents and Their Resistance to Antimicrobial Drugs. VSP 2016, 73, 1109–1115, doi:10.2298/VSP150122218D.
- Gaynes, R. The Discovery of Penicillin—New Insights After More Than 75 Years of Clinical Use. Infect. Dis. 2017, 23, 849–853, doi:10.3201/eid2305.161556.
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Opin. Microbiol. 2019, 51, 72–80, doi:10.1016/j.mib.2019.10.008.
- Dodds, D.R. Antibiotic Resistance: A Current Epilogue. Pharmacol. 2017, 134, 139–146, doi:10.1016/j.bcp.2016.12.005.
- Aminov, R. History of Antimicrobial Drug Discovery: Major Classes and Health Impact. Pharmacol. 2017, 133, 4–19, doi:10.1016/j.bcp.2016.10.001.
- de Opitz, C.L.M.; Sass, P. Tackling Antimicrobial Resistance by Exploring New Mechanisms of Antibiotic Action. Future Microbiol. 2020, 15, 703–708, doi:10.2217/fmb-2020-0048.
- Gajdács, M.; Albericio, F. Antibiotic Resistance: From the Bench to Patients. Antibiotics 2019, 8, 129, doi:10.3390/antibiotics8030129.
- Coates, A.R.M.; Hu, Y.; Holt, J.; Yeh, P. Antibiotic Combination Therapy against Resistant Bacterial Infections: Synergy, Rejuvenation and Resistance Reduction. Expert Rev. Anti Infect. Ther. 2020, 18, 5–15, doi:10.1080/14787210.2020.1705155.
- Wong, J.W.; Ip, M.; Tang, A.; Wei, V.W.; Wong, S.Y.; Riley, S.; Read, J.M.; Kwok, K.O. Prevalence and Risk Factors of Community-Associated Methicillin-Resistant Staphylococcus Aureus Carriage in Asia-Pacific Region from 2000 to 2016: A Systematic Review and Meta-Analysis. Epidemiol. 2018, 10, 1489–1501, doi:10.2147/CLEP.S160595.
- Adeiza, S.S.; Onaolapo, J.A.; Olayinka, B.O. Prevalence, Risk-Factors, and Antimicrobial Susceptibility Profile of Methicillin-Resistant Staphylococcus Aureus (MRSA) Obtained from Nares of Patients and Staff of Sokoto State-Owned Hospitals in Nigeria. GMS Hyg. Infect. Control 2020, 15, Doc25, doi:10.3205/dgkh000360.
- Queenan, K.; Häsler, B.; Rushton, J. A One Health Approach to Antimicrobial Resistance Surveillance: Is There a Business Case for It? J. Antimicrob. Agents 2016, 48, 422–427, doi:10.1016/j.ijantimicag.2016.06.014.
- Tillotson, G.S.; Zinner, S.H. Burden of Antimicrobial Resistance in an Era of Decreasing Susceptibility. Expert Rev. Anti Infect. Ther. 2017, 15, 663–676, doi:10.1080/14787210.2017.1337508.
- Heward, E.; Cullen, M.; Hobson, J. Microbiology and Antimicrobial Susceptibility of Otitis Externa: A Changing Pattern of Antimicrobial Resistance. Laryngol. Otol. 2018, 132, 314–317, doi:10.1017/S0022215118000191.
- Cillóniz, C.; Ardanuy, C.; Vila, J.; Torres, A. What Is the Clinical Relevance of Drug-Resistant Pneumococcus? Opin. Pulm. Med. 2016, 22, 227–234, doi:10.1097/MCP.0000000000000262.
- Roman, A.C.; Roman, J.V.; Flores, M.A.V.; Villaseñor, H.F.; Vidal, J.E.; Amador, S.M.; Llanos, A.M.G.; Nuñez, E.G.; Serrano, J.M.; Pastrana, G.T.; et al. Detection of Antimicrobial-Resistance Diarrheagenic Escherichia Coli Strains in Surface Water Used to Irrigate Food Products in the Northwest of Mexico. J. Food Microbiol. 2019, 304, 1–10, doi:10.1016/j.ijfoodmicro.2019.05.017.
- Relhan, N.; Pathengay, A.; Schwartz, S.G.; Flynn, H.W. Emerging Worldwide Antimicrobial Resistance, Antibiotic Stewardship and Alternative Intravitreal Agents for the Treatment of Endophthalmitis. Retina 2017, 37, 811–818, doi:10.1097/IAE.0000000000001603.
- Tomičić, Z.; Čabarkapa, I.; Čolović, R.; Đuragić, O.; Tomičić, R. Salmonella in the Feed Industry: Problems and Potential Solutions. Agron. Technol. Eng. Manag. 2019, 2, 130–137.
- Serwecińska, L.; Kiedrzyńska, E.; Kiedrzyński, M. A Catchment-Scale Assessment of the Sanitary Condition of Treated Wastewater and River Water Based on Fecal Indicators and Carbapenem-Resistant Acinetobacter Spp. Total Environ. 2021, 750, 142266, doi:10.1016/j.scitotenv.2020.142266.
- Tagliabue, A.; Rappuoli, R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Immunol. 2018, 9, 1068, doi:10.3389/fimmu.2018.01068.
- Mutairi, R.A.; Tovmasyan, A.; Haberle, I.B.; Benov, L. Sublethal Photodynamic Treatment Does Not Lead to Development of Resistance. Microbiol. 2018, 9, 1699, doi:10.3389/fmicb.2018.01699.
- Hong, J.; Hu, J.; Ke, F. Experimental Induction of Bacterial Resistance to the Antimicrobial Peptide Tachyplesin I and Investigation of the Resistance Mechanisms. Agents Chemother. 2016, 60, 6067–6075, doi:10.1128/AAC.00640-16.
- van den Bergh, B.; Michiels, J.E.; Fauvart, M.; Michiels, J. Should We Develop Screens for Multi-Drug Antibiotic Tolerance? Expert Rev. Anti Infect. Ther. 2016, 14, 613–616, doi:10.1080/14787210.2016.1194754.
- Spagnolo, F.; Rinaldi, C.; Sajorda, D.R.; Dykhuizen, D.E. Evolution of Resistance to Continuously Increasing Streptomycin Concentrations in Populations of Escherichia Coli. Agents Chemother. 2016, 60, 1336–1342, doi:10.1128/AAC.01359-15.
- Iakovides, I.C.; Kordatou, I.M.; Moreira, N.F.F.; Ribeiro, A.R.; Fernandes, T.; Pereira, M.F.R.; Nunes, O.C.; Manaia, C.M.; Silva, A.M.T.; Kassinos, D.F. Continuous Ozonation of Urban Wastewater: Removal of Antibiotics, Antibiotic-Resistant Escherichia Coli and Antibiotic Resistance Genes and Phytotoxicity. Water Res. 2019, 159, 333–347, doi:10.1016/j.watres.2019.05.025.
- Dass, S.C.; Bosilevac, J.M.; Weinroth, M.; Elowsky, C.G.; Zhou, Y.; Anandappa, A.; Wang, R. Impact of Mixed Biofilm Formation with Environmental Microorganisms on E. Coli O157:H7 Survival against Sanitization. npj Sci. Food 2020, 4, 16, doi:10.1038/s41538-020-00076-x.
- Khan, S.; Imran, A.; Malik, A.; Chaudhary, A.A.; Rub, A.; Jan, A.T.; Syed, J.B.; Rolfo, C. Bacterial Imbalance and Gut Pathologies: Association and Contribution of Coli in Inflammatory Bowel Disease. Crit. Rev. Clin. Lab. Sci. 2019, 56, 1–17, doi:10.1080/10408363.2018.1517144.
- Danson, A.E.; McStea, A.; Wang, L.; Pollitt, A.Y.; Fernandez, M.L.M.; Moraes, I.; Walsh, M.A.; MacIntyre, S.; Watson, K.A. Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia Pneumoniae’s Major Outer Membrane Protein. Biology 2020, 9, 344, doi:10.3390/biology9100344.
- Pereira, R.V.; Altier, C.; Siler, J.D.; Mann, S.; Jordan, D.; Warnick, L.D. Longitudinal Effects of Enrofloxacin or Tulathromycin Use in Preweaned Calves at High Risk of Bovine Respiratory Disease on the Shedding of Antimicrobial-Resistant Fecal Escherichia Coli. Dairy Sci. 2020, 103, 10547–10559, doi:10.3168/jds.2019-17989.
- Ellis, S.J.; Crossman, L.C.; McGrath, C.J.; Chattaway, M.A.; Hölken, J.M.; Brett, B.; Bundy, L.; Kay, G.L.; Wain, J.; Schüller, S. Identification and Characterisation of Enteroaggregative Escherichia Coli Subtypes Associated with Human Disease. Rep. 2020, 10, 7475, doi:10.1038/s41598-020-64424-3.
- Kwong, L.H.; Ercumen, A.; Pickering, A.J.; Arsenault, J.E.; Islam, M.; Parvez, S.M.; Unicomb, L.; Rahman, M.; Davis, J.; Luby, S.P. Ingestion of Fecal Bacteria along Multiple Pathways by Young Children in Rural Bangladesh Participating in a Cluster-Randomized Trial of Water, Sanitation, and Hygiene Interventions (WASH Benefits). Sci. Technol. 2020, 54, 13828–13838, doi:10.1021/acs.est.0c02606.
- Lauridsen, H.C.M.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia Coli Pathobionts Associated with Inflammatory Bowel Disease. Microbiol. Rev. 2019, 32, e00060-18, doi:10.1128/CMR.00060-18.
- Lopes, J.G.; Sourjik, V. Chemotaxis of Escherichia Coli to Major Hormones and Polyamines Present in Human Gut. ISME J. 2018, 12, 2736–2747, doi:10.1038/s41396-018-0227-5.
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Schwarz, S., Cavaco, L.M., Shen, J., Eds.; ASM Press: Washington, DC, USA, 2018; pp. 289–316, ISBN 978-1-68367-052-0.
- Card, R.M.; Cawthraw, S.A.; Garcia, J.N.; Ellis, R.J.; Kay, G.; Pallen, M.J.; Woodward, M.J.; Anjum, M.F. An In Vitro Chicken Gut Model Demonstrates Transfer of a Multidrug Resistance Plasmid from Salmonella to Commensal Escherichia Coli. mBio 2017, 8, e00777-17, doi:10.1128/mBio.00777-17.
- Ramiro, R.S.; Durão, P.; Bank, C.; Gordo, I. Low Mutational Load and High Mutation Rate Variation in Gut Commensal Bacteria. PLoS Biol. 2020, 18, e3000617, doi:10.1371/journal.pbio.3000617.
- Villa, E.B.; de la Peña, C.F.M.; Zaraín, P.L.; Cevallos, M.A.; Torres, C.; Torres, A.G.; Gracia, R.d.C.R. Comparative Genomics of a Subset of Adherent/Invasive Escherichia Coli Strains Isolated from Individuals without Inflammatory Bowel Disease. Genomics 2020, 112, 1813–1820, doi:10.1016/j.ygeno.2019.10.013.
- Papouskova, A.; Masarikova, M.; Valcek, A.; Senk, D.; Cejkova, D.; Jahodarova, E.; Cizek, A. Genomic Analysis of Escherichia Coli Strains Isolated from Diseased Chicken in the Czech Republic. BMC Vet. Res. 2020, 16, 189, doi:10.1186/s12917-020-02407-2.
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Drug Resist. 2018, 11, 1645–1658, doi:10.2147/IDR.S173867.
- Mihankhah, A.; Khoshbakht, R.; Raeisi, M.; Raeisi, V. Prevalence and Antibiotic Resistance Pattern of Bacteria Isolated from Urinary Tract Infections in Northern Iran. Res. Med. Sci. 2017, 22, 108, doi:10.4103/jrms.JRMS_889_16.
- Millan, A.S. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends Microbiol. 2018, 26, 978–985, doi:10.1016/j.tim.2018.06.007.
- Ljubojević, D.; Velhner, M.; Todorović, D.; Pajić, M.; Milanov, D. Tetracycline Resistance in Escherichia Coli Isolates from Poultry. Vet. Med. 2016, 9, 61–81.
- Puvača, N.; Lika, E.; Tufarelli, V.; Bursić, V.; Ljubojević Pelić, D.; Nikolova, N.; Petrović, A.; Prodanović, R.; Vuković, G.; Lević, J.; et al. Influence of Different Tetracycline Antimicrobial Therapy of Mycoplasma (Mycoplasma Synoviae) in Laying Hens Compared to Tea Tree Essential Oil on Table Egg Quality and Antibiotics Residues. Foods 2020, 9, 612, doi:10.3390/foods9050612.
- Abdelhalim, K.A.; Uzel, A.; Ünal, N.G. Virulence Determinants and Genetic Diversity of Adherent-Invasive Escherichia Coli (AIEC) Strains Isolated from Patients with Crohn’s Disease. Pathog. 2020, 145, 104233, doi:10.1016/j.micpath.2020.104233.
- Kilani, H.; Ferjani, S.; Mansouri, R.; Benboubaker, I.B.; Abbassi, M.S. Occurrence of Plasmid-Mediated Quinolone Resistance Determinants among Escherichia Coli Strains Isolated from Animals in Tunisia: Specific Pathovars Acquired Qnr Genes. Glob. Antimicrob. Resist. 2020, 20, 50–55, doi:10.1016/j.jgar.2019.07.023.
- Yu, Z.N.; Wang, J.; Ho, H.; Wang, Y.T.; Huang, S.N.; Han, R.W. Prevalence and Antimicrobial-Resistance Phenotypes and Genotypes of Escherichia Coli Isolated from Raw Milk Samples from Mastitis Cases in Four Regions of China. Glob. Antimicrob. Resist. 2020, 22, 94–101, doi:10.1016/j.jgar.2019.12.016.
- Farahat, E.M.; Hassuna, N.A.; Hammad, A.M.; Abdel Fattah, M.; Khairalla, A.S. Distribution of Integrons and Phylogenetic Groups among Escherichia Coli Causing Community-acquired Urinary Tract Infection in Upper Egypt. J. Microbiol. 2020, cjm-2020-0292, doi:10.1139/cjm-2020-0292.
- Ramay, B.M.; Caudell, M.A.; Rosales, C.C.; Archila, L.D.; Palmer, G.H.; Jarquin, C.; Moreno, P.; McCracken, J.P.; Rosenkrantz, L.; Amram, O.; et al. Antibiotic Use and Hygiene Interact to Influence the Distribution of Antimicrobial-Resistant Bacteria in Low-Income Communities in Guatemala. Rep. 2020, 10, 13767, doi:10.1038/s41598-020-70741-4.
- Farahani, O.; Ranjbar, R.; Jahromy, S.H.; Arabzadeh, B. Multilocus Variable-Number Tandem-Repeat Analysis for Genotyping of Escherichia Coli Strains Isolated from Hospital Wastewater, Tehran, Iran. IJPH 2020, 49, 4829, doi:10.18502/ijph.v49i12.4829.
- Riquelme, F.M.; Hernández, E.C.; Soto, M.G.; Ruiz, M.E.; Marí, J.M.N.; Fernández, J.G. Clinical Relevance of Antibiotic Susceptibility Profiles for Screening Gram-Negative Microorganisms Resistant to Beta-Lactam Antibiotics. Microorganisms 2020, 8, 1555, doi:10.3390/microorganisms8101555.
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305, doi:10.4103/joacp.JOACP_349_15.
- Rahman, S.U.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. BioMed Res. Int. 2018, 2018, 9519718, doi:10.1155/2018/9519718.
- Doma, A.O.; Popescu, R.; Mitulețu, M.; Muntean, D.; Dégi, J.; Boldea, M.V.; Radulov, I.; Dumitrescu, E.; Muselin, F.; Puvača, N.; et al. Comparative Evaluation of QnrA, QnrB, and QnrS Genes in Enterobacteriaceae Ciprofloxacin-Resistant Cases, in Swine Units and a Hospital from Western Romania. Antibiotics 2020, 9, 698, doi:10.3390/antibiotics9100698.
- Rilo, M.P.; Martín, C.-B.G.; Fernández, E.P.; Vilaró, A.; Fraile, L.; Martínez, S.M. Antimicrobial Resistance Genes in Porcine Pasteurella Multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics 2020, 9, 614, doi:10.3390/antibiotics9090614.
- Torban, A.S.;Venezia, S.N.; Kelmer, E.; Cohen, A.; Paitan, Y.; Arielly, H.; Steinman, A. Extended-Spectrum β-Lactamase-Producing Enterobacterles Shedding by Dogs and Cats Hospitalized in an Emergency and Critical Care Department of a Veterinary Teaching Hospital. Antibiotics 2020, 9, 545, doi:10.3390/antibiotics9090545.
- Falgenhauer, L.; Schwengers, O.; Schmiedel, J.; Baars, C.; Lambrecht, O.; Heß, S.; Berendonk, T.U.; Falgenhauer, J.; Chakraborty, T.; Imirzalioglu, C. Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters. Microbiol. 2019, 10, 2779, doi:10.3389/fmicb.2019.02779.
- Santos, A.C. de M.; Santos, F.F.; Silva, R.M.; Gomes, T.A.T. Diversity of Hybrid- and Hetero-Pathogenic Escherichia Coli and Their Potential Implication in More Severe Diseases. Cell Infect. Microbiol. 2020, 10, 339, doi:10.3389/fcimb.2020.00339.
- Qiu, W.; Sun, J.; Fang, M.; Luo, S.; Tian, Y.; Dong, P.; Xu, B.; Zheng, C. Occurrence of Antibiotics in the Main Rivers of Shenzhen, China: Association with Antibiotic Resistance Genes and Microbial Community. Total Environ. 2019, 653, 334–341, doi:10.1016/j.scitotenv.2018.10.398.
- Sanganyado, E.; Gwenzi, W. Antibiotic Resistance in Drinking Water Systems: Occurrence, Removal, and Human Health Risks. Total Environ. 2019, 669, 785–797, doi:10.1016/j.scitotenv.2019.03.162.
- Mölstad, S.; Löfmark, S.; Carlin, K.; Erntell, M.; Aspevall, O.; Blad, L.; Hanberger, H.; Hedin, K.; Hellman, J.; Norman, C.; et al. Lessons Learnt during 20 Years of the Swedish Strategic Programme against Antibiotic Resistance. World Health Organ. 2017, 95, 764–773, doi:10.2471/BLT.16.184374.
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Niestępski, S. The Prevalence and Characterization of Antibiotic-Resistant and Virulent Escherichia Coli Strains in the Municipal Wastewater System and Their Environmental Fate. Total Environ. 2017, 577, 367–375, doi:10.1016/j.scitotenv.2016.10.203.
- Montealegre, M.C.; Roy, S.; Böni, F.; Hossain, M.I.; Daneshmand, T.N.; Caduff, L.; Faruque, A.S.G.; Islam, M.A.; Julian, T.R. Risk Factors for Detection, Survival, and Growth of Antibiotic-Resistant and Pathogenic Escherichia Coli in Household Soils in Rural Bangladesh. Environ. Microbiol. 2018, 84, e01978-18, doi:10.1128/AEM.01978-18.
- Kaesbohrer, A.; Lebl, K.B.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in Prevalence and Characteristics of ESBL/PAmpC Producing E. Coli in Food in Germany. Microbiol. 2019, 233, 52–60, doi:10.1016/j.vetmic.2019.03.025.
- Marano, R.B.M.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Tenson, T.; Corno, G.; Kassinos, D.F.; et al. A Global Multinational Survey of Cefotaxime-Resistant Coliforms in Urban Wastewater Treatment Plants. Int. 2020, 144, 106035, doi:10.1016/j.envint.2020.106035.
- Gardy, J.L.; Loman, N.J. Towards a Genomics-Informed, Real-Time, Global Pathogen Surveillance System. Rev. Genet 2018, 19, 9–20, doi:10.1038/nrg.2017.88.
- Ferran, A.A.; Lacroix, M.Z.; Mélou, A.B.; Duhil, I.; Roques, B.B. Levers to Improve Antibiotic Treatment of Lambs via Drinking Water in Sheep Fattening Houses: The Example of the Sulfadimethoxine/Trimethoprim Combination. Antibiotics 2020, 9, 561, doi:10.3390/antibiotics9090561.
- Vilaró, A.; Novell, E.; Tarancón, V.E.; Balielles, J.; Vilalta, C.; Martinez, S.; Fraile Sauce, L.J. Antimicrobial Susceptibility Pattern of Porcine Respiratory Bacteria in Spain. Antibiotics 2020, 9, 402, doi:10.3390/antibiotics9070402.
- Mileva, R.; Karadaev, M.; Fasulkov, I.; Petkova, T.; Rusenova, N.; Vasilev, N.; Milanova, A. Oxytetracycline Pharmacokinetics After Intramuscular Administration in Cows with Clinical Metritis Associated with Trueperella Pyogenes Infection. Antibiotics 2020, 9, 392, doi:10.3390/antibiotics9070392.
- Martens, E.; Demain, A.L. The Antibiotic Resistance Crisis, with a Focus on the United States. Antibiot. 2017, 70, 520–526, doi:10.1038/ja.2017.30.
- Padmini, N.; Ajilda, A.A.K.; Sivakumar, N.; Selvakumar, G. Extended Spectrum β-Lactamase Producing Escherichia Coli and Klebsiella Pneumoniae: Critical Tools for Antibiotic Resistance Pattern. Basic Microbiol. 2017, 57, 460–470, doi:10.1002/jobm.201700008.
- Sharaha, U.; Diaz, E.R.; Riesenberg, K.; Bigio, I.J.; Huleihel, M.; Salman, A. Using Infrared Spectroscopy and Multivariate Analysis to Detect Antibiotics’ Resistant Escherichia Coli Anal. Chem. 2017, 89, 8782–8790, doi:10.1021/acs.analchem.7b01025.
- Moradigaravand, D.; Palm, M.; Farewell, A.; Mustonen, V.; Warringer, J.; Parts, L. Prediction of Antibiotic Resistance in Escherichia Coli from Large-Scale Pan-Genome Data. PLoS Comput. Biol. 2018, 14, e1006258, doi:10.1371/journal.pcbi.1006258.
- Lukačišinová, M.; Fernando, B.; Bollenbach, T. Highly Parallel Lab Evolution Reveals That Epistasis Can Curb the Evolution of Antibiotic Resistance. Commun. 2020, 11, 3105, doi:10.1038/s41467-020-16932-z.
- Swain, S.S.; Paidesetty, S.K.; Padhy, R.N. Phytochemical Conjugation as a Potential Semisynthetic Approach toward Reactive and Reuse of Obsolete Sulfonamides against Pathogenic Bacteria. Drug Dev. Res. 2020, ddr.21746, doi:10.1002/ddr.21746.
- World Health Organization. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization: Geneva, Switzerland, 2018; p. 128.
- Colson, A.R.; Megiddo, I.; Uria, G.A.; Gandra, S.; Bedford, T.; Morton, A.; Cooke, R.M.; Laxminarayan, R. Quantifying Uncertainty about Future Antimicrobial Resistance: Comparing Structured Expert Judgment and Statistical Forecasting Methods. PLoS ONE 2019, 14, e0219190, doi:10.1371/journal.pone.0219190.
- Sartelli, M.; C. Hardcastle, T.; Catena, F.; Mefire, A.C.; Coccolini, F.; Dhingra, S.; Haque, M.; Hodonou, A.; Iskandar, K.; Labricciosa, F.M.; et al. Antibiotic Use in Low and Middle-Income Countries and the Challenges of Antimicrobial Resistance in Surgery. Antibiotics 2020, 9, 497, doi:10.3390/antibiotics9080497.
- Hsia, Y.; Sharland, M.; Jackson, C.; Wong, I.C.K.; Magrini, N.; Bielicki, J.A. Consumption of Oral Antibiotic Formulations for Young Children According to the WHO Access, Watch, Reserve (AWaRe) Antibiotic Groups: An Analysis of Sales Data from 70 Middle-Income and High-Income Countries. Lancet Infect. Dis. 2019, 19, 67–75, doi:10.1016/S1473-3099(18)30547-4.
- Yan, W.; Xiao, Y.; Yan, W.; Ding, R.; Wang, S.; Zhao, F. The Effect of Bioelectrochemical Systems on Antibiotics Removal and Antibiotic Resistance Genes: A Review. Eng. J. 2019, 358, 1421–1437, doi:10.1016/j.cej.2018.10.128.
- Xie, J.; Jin, L.; He, T.; Chen, B.; Luo, X.; Feng, B.; Huang, W.; Li, J.; Fu, P.; Li, X. Bacteria and Antibiotic Resistance Genes (ARGs) in PM 5 from China: Implications for Human Exposure. Environ. Sci. Technol. 2019, 53, 963–972, doi:10.1021/acs.est.8b04630.
- Palme, J.B.; Kristiansson, E.; Larsson, D.G.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, fux053, doi:10.1093/femsre/fux053.
- Merrikh, H.; Kohli, R.M. Targeting Evolution to Inhibit Antibiotic Resistance. FEBS J. 2020, 287, 4341–4353, doi:10.1111/febs.15370.
- Marine, J.-C.; Dawson, S.-J.; Dawson, M.A. Non-Genetic Mechanisms of Therapeutic Resistance in Cancer. Rev. Cancer 2020, 20, 743–756, doi:10.1038/s41568-020-00302-4.
- Heir, E.; Møretrø, T.; Simensen, A.; Langsrud, S. Listeria Monocytogenes Strains Show Large Variations in Competitive Growth in Mixed Culture Biofilms and Suspensions with Bacteria from Food Processing Environments. J. Food Microbiol. 2018, 275, 46–55, doi:10.1016/j.ijfoodmicro.2018.03.026.
- Stubbendieck, R.M.; May, D.S.; Chevrette, M.G.; Temkin, M.I.; Pienkowski, E.W.; Cagnazzo, J.; Carlson, C.M.; Gern, J.E.; Currie, C.R. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Environ. Microbiol. 2018, 85, e02406-18, doi:10.1128/AEM.02406-18.
- de Filippis, F.; Pasolli, E.; Ercolini, D. The Food-Gut Axis: Lactic Acid Bacteria and Their Link to Food, the Gut Microbiome and Human Health. FEMS Microbiol. Rev. 2020, 44, 454–489, doi:10.1093/femsre/fuaa015.
- Iwu, C.D.; Korsten, L.; Okoh, A.I. The Incidence of Antibiotic Resistance within and beyond the Agricultural Ecosystem: A Concern for Public Health. Open 2020, 9, e1035, doi:10.1002/mbo3.1035.
- Calap, P.D.; Martínez, J.D. Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics 2018, 7, 66, doi:10.3390/antibiotics7030066.
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482–501, doi:10.3934/microbiol.2018.3.482.
- Wang, Y.; Alenazy, R.; Gu, X.; Polyak, S.W.; Zhang, P.; Sykes, M.J.; Zhang, N.; Venter, H.; Ma, S. Design and Structural Optimization of Novel 2H-Benzo[h]Chromene Derivatives That Target AcrB and Reverse Bacterial Multidrug Resistance. J. Med. Chem. 2020, 113049, doi:10.1016/j.ejmech.2020.113049.
- Impey, R.E.; Hawkins, D.A.; Sutton, J.M.; da Costa, T.P.S. Overcoming Intrinsic and Acquired Resistance Mechanisms Associated with the Cell Wall of Gram-Negative Bacteria. Antibiotics 2020, 9, 623, doi:10.3390/antibiotics9090623.
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Catena, F.; Coccolini, F.; Craig Hardcastle, T.; Roques, C.; Salameh, P. Drivers of Antibiotic Resistance Transmission in Low- and Middle-Income Countries from a “One Health” Perspective—A Review. Antibiotics 2020, 9, 372, doi:10.3390/antibiotics9070372.
- Dsani, E.; Afari, E.A.; Appiah, A.D.; Kenu, E.; Kaburi, B.B.; Egyir, B. Antimicrobial Resistance and Molecular Detection of Extended Spectrum β-Lactamase Producing Escherichia Coli Isolates from Raw Meat in Greater Accra Region, Ghana. BMC Microbiol. 2020, 20, 253, doi:10.1186/s12866-020-01935-z.
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Kassem, I.I. The Mobile Colistin Resistance Gene, Mcr-1.1, Is Carried on IncX4 Plasmids in Multidrug Resistant E. Coli Isolated from Rainbow Trout Aquaculture. Microorganisms 2020, 8, 1636, doi:10.3390/microorganisms8111636.
- Montealegre, M.C.; Rodríguez, A.T.; Roy, S.; Hossain, M.I.; Islam, M.A.; Lanza, V.F.; Julian, T.R. High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia Coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings. mSphere 2020, 5, e00704-19, doi:10.1128/mSphere.00704-19.
- 114 Alexander, J.; Hembach, N.; Schwartz, T. Evaluation of Antibiotic Resistance Dissemination by Wastewater Treatment Plant Effluents with Different Catchment Areas in Germany. Rep. 2020, 10, 8952, doi:10.1038/s41598-020-65635-4.