Video Upload Options
Neurons are structurally unique and have dendrites and axons that are vulnerable to injury. Some neurons in the peripheral nervous system (PNS) can regenerate their axons after injuries. However, most neurons in the central nervous system (CNS) fail to do so, resulting in irreversible neurological disorders. To understand the mechanisms of axon regeneration, various experimental models have been utilized in vivo and in vitro.
1. Introduction
Neurons are specialized cells with long and thin projections that are essential for neuronal functions, making connections to their targets throughout the animal body. Although these projections of different neurons generally perform similar functions such as molecular transport and transmitting signals, different types of neurons exhibit strikingly distinctive axonal morphologies. Ramon y Cajal dedicated his life to the illustration and description of neurons and neuroanatomical networks, leading to a better visual understanding—and development of the concepts—of the nervous system[1]. Cajal’s discovery was the decisive evidence for the neuron doctrine, which proposes that the nervous system is a discontinuous entity and is composed of discrete individual cells. In addition, he asked why some nervous tissues in highly evolved animals fail to regenerate after injuries that cause permanent disorders described as a ‘harsh decree’. A number of neurobiologists have discovered key mechanisms of neuronal functions and now we are even uncovering the map of the complete networks from human brains. However, the mechanisms of axon regeneration and degeneration is still under investigation with different models. Neurons are vulnerable to injury due to their projections. Some neurons in the peripheral nervous system (PNS) can regenerate their axons after injury. However, most neurons in the central nervous system (CNS) fail to regenerate, potentially resulting in irreversible neurodegeneration. Therefore, the mechanisms underlying axonal regeneration in the PNS have been extensively investigated to understand regeneration program and to find methods for promoting axon regeneration in CNS. To study axon regeneration, experimentally modeling the whole processes from injury to regeneration is useful as dissecting the complex process governed by multiple regulatory steps (Figure 1).
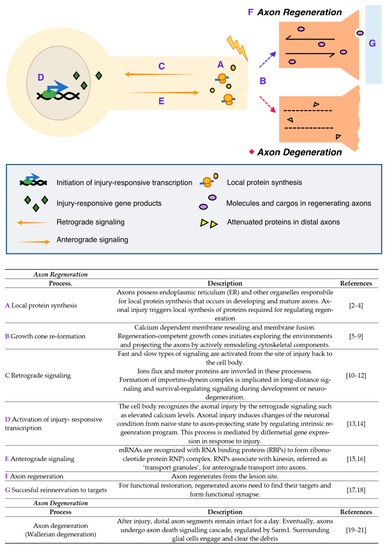
Figure 1. Injury responses in the peripheral nervous system (PNS) [2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21].
First, the axons locally synthesize multiple proteins that regulate injury-associated responses after axonal injury, which is mediated by the endoplasmic reticulum (ER) and other organelles required for protein synthesis. Simultaneously, calcium-dependent membrane resealing occurs, and the growth cone is newly constructed by cytoskeletal remodeling. Meanwhile, cell bodies recognize and activate the regeneration programs through injury-induced calcium flux and retrograde signaling that subsequently induce a shift from a naïve to a regenerative state in the neurons. Injury-responsive transcripts are produced and some of them, known as pro-regenerative transcripts or transcripts of regeneration-associated genes (RAG), bind to RNA-binding proteins (RBPs) to form ribonucleotide protein (RNP) complexes; these complexes are known to associate with kinesin and transport to the axons anterogradely. All these processes are coordinately regulated and required for successful axon regeneration as well as axonal reinnervation to the original targets, thereby resulting in the formation of functional synapses and restoring neuronal function. In contrast with regeneration, neurons also show axonal degeneration, an active process of axonal self-destruction. This is known as Wallerian degeneration[22][23][24], a phenomenon that is a characteristic feature that is only seen from neuronal cells. The distal parts of the nerve, containing the axons disconnected from their cell bodies, activate the degeneration process, resulting in the degradation of the myelin sheath and engulfment by macrophages. As briefly described here, axonal injury activates multiple biological processes in distinctive spatiotemporal regulatory systems; for example, a growth cone reforms at the tip of the transected axon, causing the regenerating axon to reinnervate its original target. Therefore, experimental models, specifically designed to investigate individual steps in the processes, are required.
Understanding differential regenerative potential from various conditions is essential for developing applications that promote axonal regeneration in the brain and spinal cord. For examples, glial proteins, such as chondroitin sulfate proteoglycan (CSPG), oligodendrocyte-myelin glycoprotein (OMgp), and myelin-associated glycoprotein (MAG), induced in response to CNS tissue injury, are absent in PNS and serve as molecular barriers that inhibit axonal regeneration[25]. This is one of the most understood key factors responsible for differential regeneration from the CNS and the PNS neurons. Interestingly, neurons in the medulla and adjacent spinal cord regenerate their injured axons into the bridges of the transplanted peripheral nerves, indicating the intrinsic regenerative potential of the CNS neurons[26]. In addition, although the PNS axons generally exhibit robust axonal regeneration, diverse factors can impair the process, thereby causing a permanent neurological deficit by either failing to reinnervate or recover the function properly[27]. These findings emphasize that both the extrinsic and intrinsic neuronal determinants should be targets for understanding the molecular mechanisms of axonal regeneration and that it would require manipulation of all the determinants for complete functional recovery following any injury [28].
To develop methods that promote functional recovery in the adult mammalian CNS and PNS, it is critical to understand the molecular mechanisms that trigger a regenerative response and the associated transcriptional regulation and epigenetic changes in injured neurons[27][29][30]. Dorsal root ganglion (DRG) neurons activate the injury-response that directs axons to either regenerate or degenerate after traumatic nerve injury. Shin et al. compared the injury-induced transcriptomic changes, i.e., differentially expressed genes (DEGs), between the regenerating and degenerating nerve segments [31]. The spatiotemporal profiles of the transcriptomes are different, indicating differential regulation between re- and degenerating neurons. However, further research is needed to determine the complexity of the mechanisms that support regeneration—and their regulation—for the development of effective therapies aimed at promoting neuronal repair in the PNS and CNS[32].
Different experimental models help us understand the mechanisms underlying axonal regeneration and identify ways to overcome technical and biological limitations. In this review, we collated results from various experimental models to comprehensively analyze the ability of neurons to regenerate in different systems. The recent innovation in the field of axon degeneration includes the identification of SARM1 as the executioner of axonal self-destruction[19][20]. The mechanisms of Wallerian degeneration and the role of nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) have been extensively dissected and Sterile Alpha and Toll Interleukin Receptor Motif-containing protein 1 (SARM1) was identified as a positive regulator of axon degeneration[20]. Osterloh et al. identified dSarm by utilizing in vivo axon degeneration assay in a loss-of-function screening in Drosophila [20]. The functional analysis of SARM1 revealed that SARM1 is an essential factor of the axon degeneration and triggers an irreversible axonal death[19][33]. Similarly, RNAi-based in vivo screening using the Drosophila models identified the new key players regulating axon degeneration such as transmembrane protein 184b (TMEM184b) and MORN repeat containing 4 (MORN4) [34][35]. In addition, an image-based large scale screening analysis identified novel functions of glycogen synthase kinase (GSK3) in axonal degeneration [36]. This loss-of-function analysis was performed at a genome-wide scale efficiently by utilizing in vitro axotomy and degeneration assay method that is introduced in this review. These innovative experimental model systems revolutionize the identifications of new players regulating axon regeneration and degeneration.
2. In Vitro Axon Injury Model
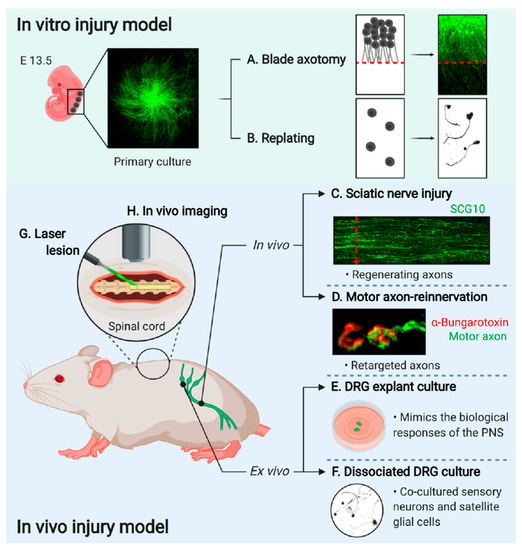
Primary cultured DRG neurons, dissected from embryonic or adult mice, are used to establish a model system for the in vitro regeneration assays and to monitor the axonal outgrowth efficiency with the manipulation of gene expression by viral vector-mediated gene delivery systems. In vitro injury can be induced by two different types of injury; axotomy (Figure 2A) or detaching and replating methods (Figure 2B).
Figure 2. Experimental models to study axon regeneration. (A) Primary cultured eDRG neurons are axotomized using a blade and the regenerating axons are investigated [13]. (B) eDRG cells are replated to investigate axonal regeneration [37]. (C) Sciatic nerve is dissected after three days of injury then immunostained with anti-SCG10 antibody to identify the regenerating axons [38]. (D) EHL muscle is observed for the target reinnervation [17]17]. (E) Adult DRGs are trypsinized and dissociated for single-cell culture[14]. (F) Adult DRGs are dissected and explanted to investigate the mechanistic pathway more easily[39]. (G) A laser lesion to the brain and spinal cord for a better understanding of the dynamics of single axons[40]. (H) In vivo imaging is performed by implantation of the spinal window in the spinal cord[41]. (Images created with BioRender.com)
2.1. Blade Axotomy
Primary cultured embryonic DRG neurons are prepared using the spot-culture method (Figure 2A). Seven days post-spot-culture, axonal injury is induced by axotomy using a surgical blade (Fine Science Tools, 10035-10), under a stereomicroscope. Axotomy terminates the delivery of all soma-derived cargoes to the distal axons, and therefore axons under the axotomy line undergo self-destructive degeneration. However, the axons that are still connected to the cell body initiates axon regeneration. These regenerating axons are quantified by counting the number of axons that cross the axotomy line within 24 to 40 h after axotomy or utilizing immunofluorescence intensities acquired by particular marker proteins such as βIII tubulin and superior cervical ganglion 10 (SCG10)[13]. Regenerating axons can be labeled with a growth-associated neuronal protein, SCG10, highly expressed in developing and regenerating axons[42][43]. Therefore, the cultures are immunostained with anti-SCG10 and anti-βIII tubulin antibodies, and regeneration indexes are calculated based on the fluorescence intensity of a square area 100 μm distal to the axotomy line and normalized to the same area 100 μm proximal to the axotomy line [13]. This assay enables to monitor the neuronal injury responses directly from the axons and the cell bodies.
2.2. Replating Assay
A replating assay can be used to monitor both axon reformation and regeneration in dispersed single neurons (Figure 2B). The dissociation of the DRGs for primary culture serves as the conditioning lesion. Replated cultured neurons mimic the pre-conditioning effect and exhibit improved neurites [44][45]. Primary cultured embryonic DRG neurons are trypsinized, dissociated into single-cell bodies, and transferred to new culture plates coated with poly-D-lysine/laminin. The replated neurons are incubated overnight at 37 °C in the presence of 5% CO2. Axonal regeneration is calculated as a measure of increasing neurite lengths from the cell body[45]. The in vitro axon injury model serves as a convenient system for evaluating the role played by specific genes in axonal regeneration under various injury conditions, providing a supportive evidence as an in vivo validation. In addition, these methods allow monitoring neuronal response without potential effects from glial cells, which is dangerous for leading to misunderstanding the true processes that naturally occur in vivo in neural tissues. Therefore, it is strongly required to remember that these experimental settings have limitations of the artifacts, which is not fully simulated in the in vivo condition.
3. In Vivo Axon Injury Model
Genetically modified animals serve as useful models for validating the targets identified based on the in vitro experiments and understanding proof-of-principle axon regeneration after spinal cord injury (Figure 2C,D,G,H) [46]. Diverse options are not available for studying genetics in peripheral sensory neurons, especially in the context of in vivo regeneration. The Advillin-Cre driver mouse line contains Cre recombinase under the regulation of the sensory neuron-specific Advillin promoter. This mouse line is a powerful tool for the targeted expression of genes of interest in sensory neurons, such as DRG neurons [47]. Additionally, the Hb9-Cre mouse is an established model for motor and interneuron-specific gene expression in the spinal cord[48][49]. As it can be used with Cre-dependent recombination to target genes from excitatory interneurons, the Hb9-Cre mouse line efficiently expresses specific genes in postmitotic motor neurons [50].
3.1. Sciatic Nerve Injury Model
3.2. Motor Axon-Reinnervation Model
The functional recovery of motor axons requires successful regeneration not only at the injury site but also at the endplates of the neuromuscular junctions for reinnervation into the target muscle [57]. The extensor hallucis longus (EHL) muscle is used to observe the reinnervation of the motor axons that regenerate through the sciatic nerves in a neuromuscular junction (Figure 2D). The EHL muscles are dissected two weeks following sciatic nerve injury to assess the reinnervation efficiency or speed of growth of motor axons in the sciatic nerve into a neuromuscular junction. Paraformaldehyde-fixed EHL muscles are stained with fluorophore-conjugated α-bungarotoxin to visualize the endplates of neuromuscular junctions. Whole-mounted or flat-mounted samples are microscopically observed to assess the extent of motor axon reinnervation [55].
3.3. Spinal Cord Injury Model
Most studies on regeneration in the CNS used retinal ganglion cells of the optic nerve or the spinal cord as the model system[58][59][60]. Relevant models used for clinical translation are contusion and hemisection injury, and these recapitulate of the pathology seen following spinal cord injuries in humans. After traumatic CNS injury, a variety of cell types, including macrophages and astrocytes, invade the injury site. These cells produce fibrotic and astrocytic scar secreting inhibitory factors that define the lesion site and serve as a physical barrier for regenerating axons[61].
Performing a laser lesion causes minimal scarring in the brain and spinal cord of mammals[41][62] (Figure 2G). When lesions are generated in axons using a highly localized laser, the extrinsic injury response is minimized[41]. Thus, the intrinsic response can be studied in more detail with the laser axotomy which can be studied for the dynamics of single axons [41]. Depending on the objective used, the power of the laser and the depth of the axon in the spinal cord determines successful lesioning[63].
3.4. In Vivo Imaging
Imaging strategies have been developed to visualize axonal regeneration in the spinal cord; these strategies allow us to investigate simple axonal dynamics after injury and axonal outgrowth after pre- and post-conditioning, retraction bulb formation, and degeneration[6][41][64]. Because wide-field microscopy does not produce images with high spatiotemporal resolution, two-photon imaging is used[63]. Two-photon imaging enables lesion development in single axons with laser pulse [41][65], exhibits deeper tissue penetration, and induces lower photo-toxicity, thereby improving the conditions required for in vivo imaging.
In vivo dynamics in the spinal cord can be visualized acutely or chronically (Figure 2H). For acute visualization, the spinal cord is stabilized with fixed holders on printed 3D microscope inserts [63]. The surgical procedure is performed precisely on a mouse placed under a two-photon microscope, all the while maintaining the physiological conditions[63]. Chronic imaging is performed at different time points after injury, making it stressful for the mouse due to the need for repetitive surgical interventions. Therefore, the spinal window has been developed to perform chronic imaging[66][67][68][69]. The implantation of the spinal windows in the spinal cord enables the imaging of the same axons for up to one year without any damage to the spinal cord [63]. Furthermore, the choice of imaging might depend on the chosen injury model.
4. Ex Vivo Injury Model
DRGs extend a single axon with two branches, one goes to the peripheral nerve and the other enters the spinal cord. After the nerve injury, the peripheral branch regenerates, while the central branch does not. Therefore, mouse DRG neurons are a valuable model for studying axon regeneration. There are two methods to culture DRG neurons to better understand neuronal functions in various experiments (Figure 2E,F).
4.1. DRG Explant Culture
Explant cultures of adult mouse DRG are used as an organotypic ex vivo injury model (Figure 2E). Once DRGs are dissected, they are plated in a culture dish pre-coated with a gelatinous protein mixture for incubation. DRGs are then gently covered with the culture medium to maintain the explants under culture conditions. The DRG explant culture is more suitable than the single-cell culture models, as it mimics the biological responses associated with the physiological and pathological conditions of the PNS [69]. The explant culture allows for the ex vivo transfer of an entire neuronal network and can be maintained for several days. Therefore, the DRG explant system offers sufficient flexibility to study various events related to biological, physiological, and pathological conditions in a cost-effective manner[69].
4.2. Dissociated DRG Neuron Culture
Culture-dissociated adult DRGs serve as another ex vivo model (Figure 2F). Primary sensory neurons and satellite glial cells can be co-cultured—from dissociated DRGs—to investigate the neuronal-glial interaction, neuritogenesis, the interaction of the axonal cone with the extracellular microenvironment, and neuronal metabolism [69]. The details of adult DRG culture have been previously described[70]. Mouse L4 and L5 DRGs are collected and incubated in DMEM supplemented with Liberase Blendzyme 3 (Roche), DNase I (Sigma), and bovine serum albumin for 15 min at 37 °C. The DRGs are then incubated with trypsin-EDTA for 15 min at 37 °C, followed by trituration. Dissociated cells are plated on culture dishes[54].
Both the DRG explants and dissociated neuronal cultures can be virally transduced through treatment with virus-containing medium and subsequent incubation at 37 °C.
References
- Santiago, P. The Croonian lecture.—La fine structure des centres nerveux. Proc. R. Soc. Lond. 1894, 55, 444–468.
- Campbell, D.S.; Holt, C.E. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 2001, 32, 1013–1026, doi:10.1016/S0896-6273(01)00551-7.
- Holt, C.E.; Schuman, E.M. The central dogma decentralized: New perspectives on RNA function and local translation in neurons. Neuron 2013, 80, 648–657, doi:10.1016/j.neuron.2013.10.036.
- Rishal, I.; Fainzilber, M. Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 2014, 15, 32–42, doi:10.1038/nrn3609.
- Ertürk, A.; Hellal, F.; Enes, J.; Bradke, F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J. Neurosci. 2007, 27, 9169–9180, doi:10.1523/JNEUROSCI.0612-07.2007.
- Ziv, N.E.; Spira, M.E. Localized and transient elevations of intracellular Ca2+ induce the dedifferentiation of axonal segments into growth cones. J. Neurosci. 1997, 17, 3568–3579, doi:10.1523/jneurosci.17-10-03568.1997.
- Bradke, F.; Fawcett, J.W.; Spira, M.E. Assembly of a new growth cone after axotomy: The precursor to axon regeneration. Nat. Rev. Neurosci. 2012, 13, 183–193, doi:10.1038/nrn3176.
- Tom, V.J.; Steinmetz, M.P.; Miller, J.H.; Doller, C.M.; Silver, J. Studies on the development and behavior of the dystrophic growth cone, the hallmark of regeneration failure, in an in vitro model of the glial scar and after spinal cord injury. J. Neurosci. 2004, 24, 6531–6539, doi:10.1523/JNEUROSCI.0994-04.2004.
- Ming, G.L.; Wong, S.T.; Henley, J.; Yuan, X.B.; Song, H.J.; Spitzer, N.C.; Poo, M.M. Adaptation in the chemotactic guidance of nerve growth cones. Nature 2002, 417, 411–418, doi:10.1038/nature745.
- Ziv, N.E.; Spira, M.E. Spatiotemporal Distribution of Ca2+ Following Axotomy and Throughout the Recovery Process of Cultured Aplysia Neurons. Eur. J. Neurosci. 1993, 5, 657–668, doi:10.1111/j.1460-9568.1993.tb00531.x.
- Ji, S.J.; Jaffrey, S.R. Axonal transcription factors: Novel regulators of growth cone- to-nucleus signaling. Dev Neurobiol. 2014, 74, 245–258, doi:10.1002/dneu.22112.
- Baleriola, J.; Walker, C.A.; Jean, Y.Y.; Crary, J.F.; Troy, C.M.; Nagy, P.L; Hengst, U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions Jimena. Cell. 2014, 158 1159–1172. doi:10.1016/j.cell.2014.07.001.
- Cho, Y.; Cavalli, V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. Embo J. 2012, 31, 3063–3078, doi:10.1038/emboj.2012.160.
- Smith, D.S.; Skene, J.H. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J. Neurosci. 1997, 17, 646–658, doi:10.1523/jneurosci.0612-07.2007.
- Shigeoka, T.; Jung, H.; Jung, J.; Turner-Bridger, B.; Ohk, J.; Lin, J.Q.; Amieux, P.S.; Holt, C.E. Dynamic Axonal Translation in Developing and Mature Visual Circuits. Cell 2016, 166, 181–192, doi:10.1016/j.cell.2016.05.029.
- Sahoo, P.K.; Smith, D.S.; Perrone-Bizzozero, N.; Twiss, J.L. Axonal mRNA transport and translation at a glance. J. Cell Sci. 2018, 131, doi:10.1242/jcs.196808.
- Magill, C.K.; Tong, A.; Kawamura, D.; Hayashi, A.; Hunter, D.A.; Parsadanian, A.; Mackinnon, S.E.; Myckatyn, T.M. Reinnervation of the tibialis anterior following sciatic nerve crush injury: A confocal microscopic study in transgenic mice. Exp. Neurol. 2007, 207, 64–74, doi:10.1016/j.expneurol.2007.05.028.
- De Lima, S.; Koriyama, Y.; Kurimoto, T.; Oliveira, J.T.; Yin, Y.; Li, Y.; Gilbert, H.Y.; Fagiolini, M.; Martinez, A.M.B.; Benowitz, L. Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9149–9154, doi:10.1073/pnas.1119449109.
- Gerdts, J.; Brace, E.J.; Sasaki, Y.; DiAntonio, A.; Milbrandt, J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Sci. (80-. ). 2015, 348, 453–457, doi:10.1126/science.1258366.
- Osterloh, J.M.; Yang, J.; Rooney, T.M.; Fox, A.N.; Adalbert, R.; Powell, E.H.; Sheehan, A.E.; Avery, M.A.; Hackett, R.; Logan, M.A.; et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Sci. (80-. ). 2012, 337, 481–484, doi:10.1126/science.1223899.
- Waller, A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Abstr. Pap. Commun. R. Soc. Lond. 1851, 5, 924–925, doi:10.1098/rspl.1843.0224.
- Coleman, M.P.; Höke, A. Programmed axon degeneration: From mouse to mechanism to medicine. Nat. Rev. Neurosci. 2020, 21, 183–196, doi:10.1038/s41583-020-0269-3.
- Wang, J.T.; Medress, Z.A.; Barres, B.A. Axon degeneration: Molecular mechanisms of a self-destruction pathway. J. Cell Biol. 2012, 196, 7–18, doi:10.1083/jcb.201108111.
- Gerdts, J.; Summers, D.W.; Milbrandt, J.; DiAntonio, A. Axon Self-Destruction: New Links among SARM1, MAPKs, and NAD+ Metabolism. Neuron 2016, 89, 449–460, doi:10.1016/j.neuron.2015.12.023.
- Yiu, G.; He, Z. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 2006, 7, 617–627, doi:10.1038/nrn1956.
- David, S.; Aguayo, A.J. Axonal elongation into peripheral nervous system “bridges” after central nervous system injury in adult rats. Sci. (80-. ). 1981, 214, 931–933, doi:10.1126/science.6171034.
- Mahar, M.; Cavalli, V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 2018, 19, 323–337, doi:10.1038/s41583-018-0001-8.
- Tedeschi, A.; Bradke, F. Spatial and temporal arrangement of neuronal intrinsic and extrinsic mechanisms controlling axon regeneration. Curr. Opin. Neurobiol. 2017, 42, 118–127, doi:10.1016/j.conb.2016.12.005.
- Lindner, R.; Puttagunta, R.; Di Giovanni, S. Epigenetic Regulation of Axon Outgrowth and Regeneration in CNS Injury: The First Steps Forward. Neurotherapeutics 2013, 10, 771–781, doi:10.1007/s13311-013-0203-8.
- Shin, J.E.; Cho, Y. Epigenetic Regulation of Axon Regeneration after Neural Injury. Mol. Cells 2017, 40, 10–16, doi:10.14348/molcells.2017.2311.
- Shin, J.E.; Ha, H.; Cho, E.H.; Kim, Y.K.; Cho, Y. Comparative analysis of the transcriptome of injured nerve segments reveals spatiotemporal responses to neural damage in mice. J. Comp. Neurol. 2018, 526, 1195–1208, doi:10.1002/cne.24404.
- Smith, T.P.; Sahoo, P.K.; Kar, A.N.; Twiss, J.L. Intra-axonal mechanisms driving axon regeneration. Brain Res. 2020, 1740, doi:10.1016/j.brainres.2020.146864.
- Osterloh, J.M.; Yang, J.; Rooney, T.M.; Fox, A.N.; Adalbert, R.; Powell, E.H.; Sheehan, A.E.; Avery, M.A.; Hackett, R.; Logan, M.A.; et al. dSarm/Sarm1 Is Required for Activation of an Injury-Induced Axon Death Pathway. Sci. (80-. ). 2012, 337, 481–484, doi:10.1126/science.1223899.
- Figley, M.D.; DiAntonio, A. The SARM1 axon degeneration pathway: Control of the NAD+ metabolome regulates axon survival in health and disease. Curr. Opin. Neurobiol. 2020, 63, 59–66, doi:10.1016/j.conb.2020.02.012.
- Bhattacharya, M.R.C.; Geisler, S.; Pittman, S.K.; Doan, R.A.; Weihl, C.C.; Milbrandt, J.; DiAntonio, A. TMEM184b Promotes Axon Degeneration and Neuromuscular Junction Maintenance. J. Neurosci. 2016, 36, 4681–4689, doi:10.1523/JNEUROSCI.2893-15.2016.
- Bhattacharya, M.R.C.; Gerdts, J.; Naylor, S.A.; Royse, E.X.; Ebstein, S.Y.; Sasaki, Y.; Milbrandt, J.; DiAntonio, A. A model of toxic neuropathy in Drosophila reveals a role for MORN4 in promoting axonal degeneration. J. Neurosci. 2012, 32, 5054–5061, doi:10.1523/JNEUROSCI.4951-11.2012.
- Gerdts, J.; Sasaki, Y.; Vohra, B.; Marasa, J.; Milbrandt, J. Image-based screening identifies novel roles for IkappaB kinase and glycogen synthase kinase 3 in axonal degeneration. J. Biol. Chem. 2011, 286, 28011—28018, doi:10.1074/jbc.m111.250472.
- Frey, E.; Valakh, V.; Karney-Grobe, S.; Shi, Y.; Milbrandt, J.; DiAntonio, A. An in vitro assay to study induction of the regenerative state in sensory neurons. Exp. Neurol. 2015, 263, 350–363, doi:10.1016/j.expneurol.2014.10.012.
- Shin, J.E.; Geisler, S.; DiAntonio, A. Dynamic regulation of SCG10 in regenerating axons after injury. Exp. Neurol. 2014, 252, 1–11, doi:10.1016/j.expneurol.2013.11.007.
- Sharthiya, H.; Seng, C.; Van Kuppevelt, T.H.; Tiwari, V.; Fornaro, M. HSV-1 interaction to 3-O-sulfated heparan sulfate in mouse-derived DRG explant and profiles of inflammatory markers during virus infection. J. Neurovirol. 2017, 23, 483–491, doi:10.1007/s13365-017-0521-4.
- Ylera, B.; Ertürk, A.; Hellal, F.; Nadrigny, F.; Hurtado, A.; Tahirovic, S.; Oudega, M.; Kirchhoff, F.; Bradke, F. Chronically CNS-Injured Adult Sensory Neurons Gain Regenerative Competence upon a Lesion of Their Peripheral Axon. Curr. Biol. 2009, 19, 930–936, doi:10.1016/j.cub.2009.04.017.
- Mason, M.R.J.; Lieberman, A.R.; Grenningloh, G.; Anderson, P.N. Transcriptional upregulation of SCG10 and CAP-23 is correlated with regeneration of the axons of peripheral and central neurons in vivo. Mol. Cell. Neurosci. 2002, 20, 595–615, doi:10.1006/mcne.2002.1140.
- Voria, I.; Hauser, J.; Axis, A.; Schenker, M.; Bichet, S.; Kuntzer, T.; Grenningloh, G.; Barakat-Walter, I. Improved sciatic nerve regeneration by local thyroid hormone treatment in adult rat is accompanied by increased expression of SCG10. Exp. Neurol. 2006, 197, 258–267, doi:10.1016/j.expneurol.2005.10.001.
- Saijilafu; Hur, E.M.; Liu, C.M.; Jiao, Z.; Xu, W.L.; Zhou, F.Q. PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun. 2013, 4, doi:10.1038/ncomms3690.
- Zou, H.; Ho, C.; Wong, K.; Tessier-Lavigne, M. Axotomy-induced smad1 activation promotes axonal growth in adult sensory neurons. J. Neurosci. 2009, 29, 7116–7123, doi:10.1523/JNEUROSCI.5397-08.2009.
- Lee, J.K.; Zheng, B. Axon regeneration after spinal cord injury: Insight from genetically modified mouse models. Restor. Neurol. Neurosci. 2008, 26, 175–182.
- Zurborg, S.; Piszczek, A.; Martínez, C.; Hublitz, P.; Al Banchaabouchi, M.; Moreira, P.; Perlas, E.; Heppenstall, P.A. Generation and characterization of an Advillin-Cre driver mouse line. Mol. Pain 2011, 7, 1–10, doi:10.1186/1744-8069-7-66.
- Wilson, J.M.; Hartley, R.; Maxwell, D.J.; Todd, A.J.; Lieberam, I.; Kaltschmidt, J.A.; Yoshida, Y.; Jessell, T.M.; Brownstone, R.M. Conditional rhythmicity of ventral spinal interneurons defined by expression of the Hb9 homeodomain protein. J. Neurosci. 2005, 25, 5710–5719, doi:10.1523/JNEUROSCI.0274-05.2005.
- Arber, S.; Han, B.; Mendelsohn, M.; Smith, M.; Jessell, T.M.; Sockanathan, S. Requirement for the Homeobox Gene Hb9 in the Consolidation of Motor Neuron Identity. Neuron 1999, 23, 659–674, doi:10.1016/S0896-6273(01)80026-X.
- Caldeira, V.; Dougherty, K.J.; Borgius, L.; Kiehn, O. Spinal Hb9::Cre-derived excitatory interneurons contribute to rhythm generation in the mouse. Sci. Rep. 2017, 7, 1–12, doi:10.1038/srep41369.
- Lee, J.; Shin, J.E.; Lee, B.; Kim, H.; Jeon, Y.; Ahn, S.H.; Chi, S.W.; Cho, Y. The stem cell marker Prom1 promotes axon regeneration by down-regulating cholesterol synthesis via Smad signaling. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 15955–15966, doi:10.1073/pnas.1920829117.
- Richardson, P.M.; Ebendal, T. Nerve growth activities in rat peripheral nerve. Brain Res. 1982, 246, 57–64, doi:10.1016/0006-8993(82)90141-X.
- Neumann, S.; Woolf, C.J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 1999, 23, 83–91, doi:10.1016/S0896-6273(00)80755-2.
- Abe, N.; Cavalli, V. Nerve injury signaling. Curr. Opin. Neurobiol. 2008, 18, 276–283, doi:10.1016/j.conb.2008.06.005.
- Shin, J.E.; Cho, Y.; Beirowski, B.; Milbrandt, J.; Cavalli, V.; DiAntonio, A. Dual Leucine Zipper Kinase Is Required for Retrograde Injury Signaling and Axonal Regeneration. Neuron 2012, 74, 1015–1022, doi:10.1016/j.neuron.2012.04.028.
- Hanz, S.; Fainzilber, M. Retrograde signaling in injured nerve-The axon reaction revisited. J. Neurochem. 2006, 99, 13–19, doi:10.1111/j.1471-4159.2006.04089.x.
- Cavalli, V.; Kujala, P.; Klumperman, J.; Goldstein, L.S.B. Sunday driver links axonal transport to damage signaling. J. Cell Biol. 2005, 168, 775–787, doi:10.1083/jcb.200410136.
- Hilton, B.J.; Assinck, P.; Duncan, G.J.; Lu, D.; Lo, S.; Tetzlaff, W. Dorsolateral funiculus lesioning of the mouse cervical spinal cord at C4 but not at C6 results in sustained forelimb motor deficits. J. Neurotrauma 2013, 30, 1070–1083, doi:10.1089/neu.2012.2734.
- Sun, F.; Park, K.K.; Belin, S.; Wang, D.; Lu, T.; Chen, G.; Zhang, K.; Yeung, C.; Feng, G.; Yankner, B.A.; et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 2011, 480, 372–375, doi:10.1038/nature10594.
- Tuszynski, M.H.; Steward, O. Concepts and Methods for the Study of Axonal Regeneration in the CNS. Neuron 2012, 74, 777–791, doi:10.1016/j.neuron.2012.05.006.
- Cregg, J.M.; DePaul, M.A.; Filous, A.R.; Lang, B.T.; Tran, A.; Silver, J. Functional regeneration beyond the glial scar. Exp. Neurol. 2014, 253, 197–207, doi:10.1016/j.expneurol.2013.12.024.
- Canty, A.J.; Teles-Grilo ruivo, L.M.; Nesarajah, C.; Song, S.; Jackson, J.S.; Little, G.E.; De Paola, V. Synaptic elimination and protection after minimal injury depend on cell type and their prelesion structural dynamics in the adult cerebral cortex. J. Neurosci. 2013, 33, 10374–10383, doi:10.1523/JNEUROSCI.0254-13.2013.
- Schaffran, B.; Hilton, B.J.; Bradke, F. Imaging in vivo dynamics of sensory axon responses to CNS injury. Exp. Neurol. 2019, 317, 110–118, doi:10.1016/j.expneurol.2019.02.010.
- Kerschensteiner, M.; Schwab, M.E.; Lichtman, J.W.; Misgeld, T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat. Med. 2005, 11, 572–577, doi:10.1038/nm1229.
- Lorenzana, A.O.; Lee, J.K.; Mui, M.; Chang, A.; Zheng, B. A Surviving Intact Branch Stabilizes Remaining Axon Architecture after Injury as Revealed by InVivo Imaging in the Mouse Spinal Cord. Neuron 2015, 86, 947–954, doi:10.1016/j.neuron.2015.03.061.
- Farrar, M.J.; Bernstein, I.M.; Schlafer, D.H.; Cleland, T.A.; Fetcho, J.R.; Schaffer, C.B. Chronic in vivo imaging in the mouse spinal cord using an implanted chamber. Nat. Methods 2012, 9, 297–302, doi:10.1038/nmeth.1856.
- Fenrich, K.K.; Weber, P.; Hocine, M.; Zalc, M.; Rougon, G.; Debarbieux, F. Long-term in vivo imaging of normal and pathological mouse spinal cord with subcellular resolution using implanted glass windows. J. Physiol. 2012, 590, 3665–3675, doi:10.1113/jphysiol.2012.230532.
- Figley, S.A.; Chen, Y.; Maeda, A.; Conroy, L.; McMullen, J.D.; Silver, J.I.; Stapleton, S.; Vitkin, A.; Lindsay, P.; Burrell, K.; et al. A Spinal Cord Window Chamber Model for In Vivo Longitudinal Multimodal Optical and Acoustic Imaging in a Murine Model. Plos One 2013, 8, doi:10.1371/journal.pone.0058081.
- Tedeschi, A.; Dupraz, S.; Laskowski, C.J.; Xue, J.; Ulas, T.; Beyer, M.; Schultze, J.L.; Bradke, F. The Calcium Channel Subunit Alpha2delta2 Suppresses Axon Regeneration in the Adult CNS. Neuron 2016, 92, 419–434, doi:10.1016/j.neuron.2016.09.026.
- Fornaro, M.; Sharthiya, H.; Tiwari, V. Adult mouse DRG explant and dissociated cell models to investigate neuroplasticity and responses to environmental insults including viral infection. J. Vis. Exp. 2018, 2018, 1–8, doi:10.3791/56757.
- Santiago, P. The Croonian lecture.—La fine structure des centres nerveux. Proc. R. Soc. Lond. 1894, 55, 444–468.
- Fornaro, M.; Sharthiya, H.; Tiwari, V. Adult mouse DRG explant and dissociated cell models to investigate neuroplasticity and responses to environmental insults including viral infection. J. Vis. Exp. 2018, 2018, 1–8, doi:10.3791/56757.
- Fornaro, M.; Sharthiya, H.; Tiwari, V. Adult mouse DRG explant and dissociated cell models to investigate neuroplasticity and responses to environmental insults including viral infection. J. Vis. Exp. 2018, 2018, 1–8, doi:10.3791/56757.