
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Peter Surai | + 3815 word(s) | 3815 | 2021-02-01 09:22:51 | | | |
| 2 | Vivi Li | Meta information modification | 3815 | 2021-02-01 11:01:38 | | | | |
| 3 | Vivi Li | Meta information modification | 3815 | 2021-02-01 11:03:55 | | | | |
| 4 | Vivi Li | + 3 word(s) | 3818 | 2021-02-02 02:28:21 | | |
Video Upload Options
Redox biology is a very quickly developing area of modern biological sciences, and roles of redox homeostasis in health and disease have recently received tremendous attention. There are a range of redox pairs in the cells/tissues responsible for redox homeostasis maintenance/regulation. In general, all redox elements are interconnected and regulated by various means, including antioxidant and vitagene networks. The redox status is responsible for maintenance of cell signaling and cell stress adaptation. Physiological roles of redox homeostasis maintenance in avian species, including poultry, have received limited attention and are poorly characterized. However, for the last 5 years, this topic attracted much attention, and a range of publications covered some related aspects. In fact, transcription factor Nrf2 was shown to be a master regulator of antioxidant defenses via activation of various vitagenes and other protective molecules to maintain redox homeostasis in cells/tissues. It was shown that Nrf2 is closely related to another transcription factor, namely, NF-κB, responsible for control of inflammation; however, its roles in poultry have not yet been characterized. Therefore, the aim of this study is to describe a current view on NF-κB functioning in poultry with a specific emphasis to its nutritional modulation under various stress conditions.
1. Introduction
Redox biology is a very quickly developing area of modern biological sciences, and roles of redox homeostasis in health and disease have recently received tremendous attention [1][2][3][4][5][6]. There are a range of redox pairs in cells/tissues responsible for redox homeostasis maintenance/regulation. They include, but are not limited to, NAD+/NADH, NADP+/NADPH, GSSH/GSH (glutathione system), Trxox/Trxred (thioredoxin system), protein thiolsox/protein thiolsred. It is believed that redox signaling is tightly integrated with various homeostatic mechanisms [7] and all redox elements are interconnected and regulated by various means, including antioxidant and vitagene networks [1]. The redox status is responsible for maintenance of cell signaling and cell stress adaptation. There are a range of redox sensors which determine redox imbalance and activate various pathways for its re-establishment. Among them are proteins Keap1, an inhibitor of Nrf2, and IκB, an inhibitor of NF-κB, which have received a lot of recent attention. Indeed, oxidation of SH groups in Cys of Keap1 or phosphorylation of IκB are important triggers for nuclear translocation and activation of Nrf2 and NF-κB—important players in the redox homeostasis regulation [6]. In particular, a recent model suggests regulation of all collaborating metabolic organs in the body through changes in circulating redox metabolites [5].
The physiological roles of redox homeostasis maintenance in avian species, including poultry, are poorly characterized. However, for the last 5 years, this topic attracted a lot of attention, and a range of publications covered some related aspects. Indeed, the redox system imbalance is shown to be associated with protein oxidation and impaired quality of poultry meat [8][9]. In broilers, subjected to dietary and heat stress, magnesium supplementation was indicated to improve redox status and meat quality [10]. The influence of selenium and selenoproteins in maintaining redox balance and immune responses of poultry and pigs was presented [11], and the effect of oxidative stress and redox disbalance on inflammation, including a detailed immune system investigation, was discussed [12][13]. Oxidative stress-related disturbances of the redox balance in the poultry gut have also been described [13][14][15][16]. The long-term effects of Ochratoxin A on the glutathione redox system in chickens have been investigated [17], and the protective effects of milk thistle on redox-homeostasis imbalance of duck liver imposed by mycotoxins [18] were shown. Furthermore, the detrimental effects of heavy metals (e.g., As) on redox imbalance in chickens have been reported [19]. Nutritional modulation of the antioxidant capacities and redox homeostasis in poultry by selenium [13][20], vitamin E [21], and carotenoids [22], including astaxanthin [23], has been described. Recently, the vitagene concept of stress adaptation was developed, and questions related to redox balance maintenance in poultry under various stress conditions were addressed [1]. In fact, the vitagene family includes superoxide dismutase (SOD), heat shock protein 70 (HSP70), heme oxygenase 1(HO-1), elements of thioredoxin and glutathione systems, and sirtuins [24][25][26]. Indeed, induction/activation of the aforementioned genes leading to synthesis/expression of protective molecules helps animals/poultry adapt to stress by using their internal resources to the maximum extent.
2. NF-κB and Oxidative Stress
Free-radical production is considered to be an important process in biological systems responsible for the antibacterial action of oxidative burst in phagocytes, cell signaling, and stress adaptation [7]. However, an excess of reactive oxygen and nitrogen species (RONS) due to high level of stress or a compromised antioxidant system leads to damages to major biological molecules (proteins, polyunsaturated fatty acids (PUFAs), DNA, etc.) associated with immunosuppression, gut health problems, and decreased productive and reproductive performance of poultry [27]. Therefore, a variety of protective mechanisms have been developed during evolution to deal with RONS excess, and many transcription factors are involved in this process via regulating vitagenes and a myriad of antioxidant enzymes in stress conditions [1].
There are a range of transcription factors acting cooperatively with NF-κB. For-example, NF-κB and STAT3 are shown to regulate common pathways and share regulatory binding sites of various protective genes, while HIF and NF-κB are reported to share common activating stimuli, regulators, and targets [28]. Indeed, the redox balance is believed to be orchestrated by a range of transcription factors, including Nrf2, NF-κB, activator protein 1 (AP-1), FoxO, peroxisome proliferator-activated receptors (PPARs), peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α), p53, and mitogen-activated protein kinase (MAPK; Figure 3 [29][30]). It seems likely that transcription factors and vitagenes are involved in the regulation of redox status by effectively modulating the expression and activity of ROS-generating enzymes and antioxidant enzymes [31].
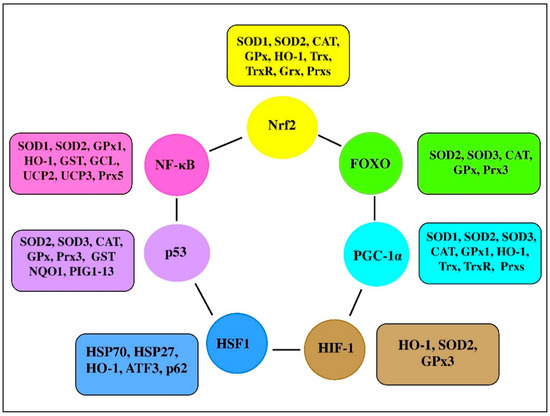
Figure 3. Transcription factors and their clients involved in redox homeostasis regulation (adapted from [1][30][31][32]).
NF-κB has long been considered to be a prototypical proinflammatory signaling pathway stimulating the immune system in response to various stimuli, including physical, physiological, and/or oxidative stress. For example, NF-κB is a key target in receptor-independent hypothalamic microinflammation [33] associated with intracellular organelle stress, including RNA stress response [34], endoplasmic reticulum (ER) stress [35], and defective autophagy [36]. NF-κB is involved in the regulation of many important physiological processes; however, its overactivation has been shown to be associated with increased risk of disease, while NF-κB suppression is associated with risk reduction [37]. Taking the former into account, understanding the role of NF-κB signaling in stress adaptation awaits further investigation. For example, HO-1 can improve cell protection from apoptosis by stimulating free heme catabolism. Interestingly, the HO-1 promoter region contains an NF-κB responsive element and, therefore, HO-1 expression is regulated by NF-κB, as well as by other transcription factors [38]. A central role for NF-κB in regulating mitochondrial respiration has been suggested [39]. In fact, by controlling the balance between glycolysis and respiration for energy provision, NF-κB is involved in energy homeostasis and metabolic adaptation [40]. The authors suggested to consider NF-κB as an important checkpoint connecting cell activation and proliferation with energy sensing and metabolic homeostasis. Since mitochondria are the main ROS source in the cell, it could be that NF-κB signaling is involved in the regulation of ROS formation, detoxification, and the maintenance of redox homeostasis.
3. NF-κB in Poultry Production
The regulatory roles of NF-κB in poultry are still poorly understood, but accumulating information clearly indicates that, similar to mammals, NF-κB is a main regulator of many important processes, including inflammation in avian species. In 1993, complementary DNA (cDNA) clones encoding the chicken NF-κB p65 subunit were isolated, and, according to the information provided by the authors, chicken NF-κB can be briefly characterized as follows [41]:
-
Chicken p65 was shown to be approximately 55% identical to the mouse and human p65 proteins. Similar to its mammalian counterpart, chicken p65 contains the Rel homology domain (RHD) in its N-terminal consisting of 286 amino acids and the putative transactivation domain in its C-terminal region;
-
It was proven that the RHD was highly conserved between the chicken and mammalian p65 proteins;
-
The highest expression of a 2.6 kb transcript of p65 was detected in the spleen. It was also detected in other organs;
-
A fusion protein containing the RHD of chicken p65 was reported to bind to a consensus kappa B-site;
-
p65 was shown to form one or more complexes with various cellular proteins, including p50, p105, and c-Rel in chicken spleen cells [41].
Furthermore, the cDNA clones encoding chicken p50B/p97 were isolated [42]. The amino-acid sequence of the precursor protein p97 was found to be characterized by a conserved structure. In particular, it was shown to have 86% identity in the RHD and lower (56%) identity in the ankyrin repeat domain (ARD) to human p50B/p97. Similar to previous findings, expression of this gene was also found to be highest in the chicken spleen [42]. In 1995 from a chicken genomic library, a clone containing the avian I kappa B-alpha gene was isolated [43]. Main characteristics of I kappa B-alpha can be summarized as follows: recognizable promoter elements (i.e., TATA and CAAT boxes) were not found in avian I kappa B-α. There were seven putative Rel/NF-kappa B binding sites in avian I kappa B-α. When transfected into cells which produce I kappa B-α, a CAT reporter construct containing the 5′ upstream region of I kappa B-α was expressed. The regulatory elements promoting I kappa B-α expression were identified within 1000 nt of the transcription start site. I kappa B-alpha was shown to be found as a single-copy gene per haploid genome. This gene was expressed in avian hematopoietic tissues and in lymphoid cells transformed by avian reticuloendotheliosis virus [43]. It was suggested that, similar to mammals, in chicken, p65 and c-Rel comprise components of the protein complexes that are able to bind to the kappa B-like sequence. This binding could lead to the progressively activated expression of the chicken lysozyme gene observed during the terminal differentiation of macrophages [44].
In 2001, Piffat et al. constructed and characterized a composite cDNA encoding most of the chicken RelB transcription factors [45], and their results can be summarized as follows: within the RH domain, chicken RelB (cRelB) protein was characterized by a high degree of sequence similarity to other vertebrate RelB proteins. However, outside this domain, cRelB was substantially less conserved. cRelB was found to be more widely expressed than mammalian RelB, and it was identified to have functional properties similar to other vertebrate RelB proteins. cRelB was reported to be unable to bind DNA in a homodimer form; however, it could form DNA-binding heterodimers with NF-kappaB p50 or p52. Overexpressed cRelB was shown to be present in the nucleus in chicken embryo fibroblasts. The nonconserved C-terminal sequences of cRelB contained a transactivation domain found in chicken and mouse fibroblasts [45]. A new isoform of chicken myeloid differentiation factor 88 (MyD88-2) expression was detected in a range of tissues tested and its overexpression was found to significantly induce the activation of NF-κB in vitro [46]. Recently the duck IKKα (duIKKα) gene was cloned and characterized. In fact, DuIKKα was reported to encode a protein containing 757 amino acids and having high sequence identities with the goose IKKα. Duck liver and heart were characterized by a high expression of duIKKα messenger RNA (mRNA), while its expression was reported in all tested tissues, including muscular stomach, spleen, heart, liver, lung, kidney, cerebellum, cerebrum, windpipe, muscle, glandular stomach, thymus, duodenum, cecum, pancreas, and bursa of Fabricius [47]. An important role of du IKKα in NF-κB regulation has been demonstrated by increasing or inhibiting expression of duIKKα. On the one hand, overexpression of duIKKα was shown to substantially increase NF-κB activity with subsequent induction of cytokines interferon beta (IFN-β), IL-1β, IL-6, and IL-8 in duck embryo fibroblasts. On the other hand, knockdown of duIKKα was found to significantly decrease LPS-, poly(I:C)-, poly(dA:dT)-, duck enteritis virus (DEV)-, or duck Tembusu virus (DTMUV)-induced NF-κB activation [47]. It seems likely that IKKα is evolutionarily conserved. In fact, phosphorylation of Ser176 and Ser180 in the active center of IKKα is believed to be vital to IKKα activation, and those Ser residues were shown to be well conserved among mammals, birds, and fish [47].
It was shown that the NF-κB family of transcription factors contribute to activation-induced cytidine deaminase-mediated gene conversion in chickens [48]. Gallus heat-shock cognate protein 70 was shown to regulate RelA/p65 gene expression induced by Apoptin, a nonstructural protein of chicken anemia virus [49]. In chicken heterophils, bacterial TLR agonists were indicated to activate NF-κB-mediated leukotriene B4 and prostaglandin E2 production [50]. A switchlike response in NF-κB activity is based on the existence of a threshold in the NF-κB signaling module, and phosphorylation of the Ser-578 residue of the scaffolding protein caspase recruitment domain (CARD)-containing protein 1 (CARMA1) was shown to account for the feedback [51]. It is known that tumor necrosis factor receptor-associated factors (TRAFs) are responsible for activation of various signaling cascades, being key regulatory proteins in NF-κB signaling pathways [52]. It seems likely that avian TRAFs play important roles in defending against both RNA and DNA virus infection. In fact, chicken TRAF3 (chTRAF3) was shown to encode a protein of 567 amino acids with high identity to TRAF3 homologs from mammals being abundantly expressed in the spleen, thymus, lung, and small intestine [53]. Of note, the authors showed that Newcastle disease virus F48E9 challenge was responsible for TRAF3 suppression in chicken embryo fibroblast cells. Recently, the full-length duck TRAF6 (duTRAF6) cDNA from embryo fibroblasts was cloned, and it was shown that duTRAF6 was widely expressed in different tissues. Interestingly, overexpression of duTRAF6 was found to activate NF-κB and induce interferon-β expression [54]. It has been shown that goose TRAF6 shared similar features with the TRAF6 of other avian species, being an essential regulator for inducing the activity of NF-κB and playing important roles in innate immune response [55]. The amino-acid sequence of pigeon FRAF6 (piTRAF6) was shown to share a strong identity with that of other birds. Furthermore, piTRAF6 expression was shown in all examined tissues, including heart, lung, spleen, thigh muscle, large intestine, caecum, kidney small intestine, brain, bursa of Fabricius, rib, and muscular stomach [56]. The heart was characterized by the highest level of piTRAF6 transcript, and the muscular stomach had the lowest level of piTAF6 transcript. On the one hand, overexpression of piTRAF6 was shown to induce NF-κB in a dose-dependent manner with increased IFN-β expression. On the other hand, piTRAF6 knockdown was reported to suppress NF-κB activation in HEK293T cells [56]. Furthermore, the pigeon TRAF3 (PiTRAF3) gene was reported to be highly expressed in the spleen, lung, kidney, brain, thymus, and muscle, while a moderate expression was observed in the small and large intestines, with relatively weak expression in the heart and liver [57].
Among the five major families of pattern recognition receptors (PRRs), Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), in particular, NOD1, recently received major attention in relation to their roles in avian immunity via NF-κB regulation. Indeed, NF-κB is considered to be the major transcription factor involved downstream of the TLR signaling pathway [58]. Avian TLRs are shown to be different from their mammalian counterparts: absence of TLR8 and TLR9, along with presence of TLR1La, TLR1Lb, TLR15, and TLR21 [59]. Therefore, in chickens, 10 TLR receptor genes were identified: TLR1LA, TLR1LB, TLR2B, TLR2A, TLR3, TLR4, TLR5, TLR7, TLR15 [60], and TLR21 [61]. Among them, TLR1LA, TLR1LB, TLR2A, TLR2B, TLR4, TLR5, and TLR15 are responsible for bacterial component (lipoproteins, peptidoglycans, LPS, and flagellin) detection, while TLR3 and TLR7 detect viruses (double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), imidazoquinoline compounds), and TRL21 detects CpG oligodeoxynucleotides in viruses and bacteria [61]. Initially, it was reported that chicken TLR2 and TLR4 can mediate LPS-stimulated oxidative burst, while CD14 and TLR2 are involved in the mediation of lipoteichoic acid-stimulated oxidative burst in heterophils [62]. The tissue-specific expression of chicken TLRs (TLR2A, TLR3, TLR4, TLR5, TLR7, TLR15, and TLR21) during embryonic development was evaluated and early (third embryonic day) expression of all the TLR mRNAs was reported [63]. Furthermore, TLR1 (type 1 and 2), TLR2 (type 1 and 2), and TLRs 3–5, 7, 15, and 21 were shown to be expressed in the chicken follicular theca. The connection of the TLRs to NF-κB was proven experimentally; the expression of IL-1β, IL-6, chemotactic and angiogenic factor (CXCLi2), and IFN-β in tissues incubated with LPS was downregulated by an inhibitor of NF-κB [58].
It seems likely that NF-κB is involved in the activation of avian antimicrobial peptides. For example, chicken intestine defensins (e.g., AvBD13) were suggested to be endogenous ligands for TLR4 able to enhance the proliferation of monocytes via the NF-κB pathway [64]. It should be mentioned that cathelicidins (CATHs), short cationic host defense peptides, also act in close concert with NF-κB. Indeed, in macrophages primed by LPS, pigeon CATH2 was shown to act through MAPK and NF-κB signaling pathways to enhance expression of the anti-inflammatory cytokine, while downregulating the expressions of inducible nitric oxide synthase and proinflammatory cytokines and inhibiting the TLR4 pathway [65]. Furthermore, NK-lysin/granulysin (NKL), an antimicrobial cationic peptide expressed in natural killer cells and cytotoxic T lymphocytes, was identified in different avian species, including chicken, turkey, zebra finch, and quail, and the 5′ flanking region of quail NKL was shown to contain two NF-κB-binding sites [66], suggesting participation of NF-κB in regulation of NKL activity.
In hen vaginal cells, NF-κB was shown to be the transcription factor responsible for the expression of various proinflammatory cytokines and chemokines. In fact, in response to the ligands of TLR3, 4, and 21, increased expression of IL1B, IL6, and CXCLi2 was observed, while IL1B expression was found in response to the ligands of TLR5 and 7 [67]. The authors suggested that NF-κB-dependent expression of cytokines might provide the important defense capability of vaginal tissue to bacterial and viral infections. Activation of TLR3 was shown to induce the expression of NF-κB and the production of type-I interferon [68]. IFN-κ (a type I IFN) in both chicken and duck was found to be constitutively expressed in a range of tissues, including spleen, skin, lung, and peripheral blood mononuclear cells (PBMCs), and it could be induced after treatment with virus in PBMCs [69]. The duck TLR4 (duTLR4) gene was shown to be strongly expressed in the liver, kidney, spleen, intestine, and brain [70].
Goose TLR3 was shown to be analogous to mammalian TLR3 and recognized double-stranded RNA with subsequent activation of NF-κB [68]. In fact, the goose TLR3 gene was shown to encode a protein containing 896 amino acids, sharing 46.7–84.4% homology with other species with highest expression in the pancreas and lowest in the skin. The authors showed that geese infected with H5N1 were characterized by significant upregulation of TLR3 in various tissues, including the lung and brain [68]. The goose TLR5 (gTLR5) gene was shown to be expressed in all studied tissues, including high expression in the liver, spleen, and brain, moderate expression in kidney, lung, heart, bone marrow, small intestine, large intestine, and PBMCs, and minimal expression in the cecum [71]. It was also shown that gTLR5 can detect flagellin from Salmonella Typhimurium with subsequent NF-κB activation in HEK293 cells. It seems likely that there is a tissue-specific regulation of TLR expression in the process of orchestrating the immune response against bacterial pathogens [71]. Goose TLR2-1 was also shown to play an important role in the recognition of Mycoplasma fermentans lipopeptide, Mycoplasma gallisepticum (MG) and Salmonella enteritidis (SE), and it induced the activation of NF-κB [72]. Furthermore, in HEK293T cells, flagellin was shown to induce pigeon NF-κB via TLR5 activation. This was associated with significant upregulation of IL-1β, IL-8, TNF-α, and IFN-γ. Importantly, the levels of TLR5, NF-κB, IL-6, IL-8, chemokine ligand 5 (CCL5), and IFN-γ mRNA were significantly upregulated as a result of flagellin stimulation of pigeon splenic lymphocytes. As could be expected, goose TLR5 knockdown was shown to be associated with the significantly downregulated expression of NF-κB and related cytokines/chemokines [73]. Interestingly, the antiviral activity of pigeon IFN-α is believed to depend on the expression of NF-κB [74]. It is known that single-stranded viral RNAs and antiviral imidazoquinoline compounds can be recognized by TLR7 with subsequent NF-κB activation. Recently, it was shown that, in pigeon, agonist R848 (imidazoquinoline) can activate NF-κB via TLR7 [75].
It seems likely that chicken NOD1 activation in response to pathogenic invasion is of great importance for immune defense. In partridge chicken, NOD1 was shown to be widely distributed in various tissues, with the highest expression found in testes. Of note, as a result of S. enterica serovar Enteritidis infection, induced expression of chNOD1, as well as the effector molecule NF-κB, was observed in the spleen tissue [76]. Duck NOD1 (duNOD1) was shown to be widely distributed in various organs, including heart, liver, spleen, lung, kidney, cerebrum, cerebellum, colon, glandular stomach, thymus, and bursa of Fabricius tissue with the highest expression found in the liver. Of note, duNOD1 overexpression induced NF-κB, TNF-α, and IL-6 activation in duck embryo fibroblasts (DEFs), while silencing duNOD1 was indicated to decrease the activity of NF-κB in stimulated DEFs [77].
Chicken IL-26 was shown to regulate immune responses through the NF-κB and the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) Janus kinase signaling pathways [78]. Similarly, chicken IL-11 was shown to bind to IL-11R and activated the NF-κB, JAK/STAT, and MAPK signaling pathways, leading to modulation of T helper 1 (Th1)/Th17 and Th2 cytokine production in chicken cell lines [79]. Chicken interleukin-17B was shown to induce the NF-κB signaling pathway, leading to increased expression of proinflammatory cytokines playing a critical role in host defense against the bacterial pathogens [80]. In eukaryotic and prokaryotic expression systems, recombinant chicken TNF-α was generated to demonstrate its biological activity. In particular, as a result of binding to TNF-α receptor 1, the cytokine was shown to induce a complex signaling cascade leading to induction of the classical NF-κB pathway [81].
In Gaoyou duck skeletal muscle (Anas platyrhynchos domesticus), NF-κB motifs (binding sites) were identified, which are believed to be responsible for transcriptional regulation of the slow skeletal muscle troponin I (TNNI1) gene [82]. It seems likely that chicken NF-κB plays a central role in antiviral defense. In fact, chicken tracheal epithelial cells were shown to initiate effective antiviral responses after stimulation with TLR ligands as a result of interferon regulatory factor 7 (IRF7) and NF-κB signaling pathways associated with activation of other cells, such as macrophages [83].
Receptor activator of NF-κB ligand (RANKL), a new member of the chicken TNF superfamily, was recently identified and characterized [60]. Therefore, chicken RANKL (chRANKL), sharing ~59–62% identity with mammalian RANKL, was shown to be ubiquitously expressed in chicken tissues. In nonlymphoid tissues, chRANKL mRNA expression levels were shown to be highest in muscle, while, in lymphoid tissues, the highest RANKL expression was found to be in the thymus, followed by the upper gut and the bone marrow [84]. Recently identified and functionally characterized chicken leukocyte immunoglobulin-like receptor A5 (LILRA5) was reported to activate/induce NF-κB, as well as other immunoregulatory pathways [85].
References
- Surai, P.F. Vitagenes in Avian Biology and Poultry Health; Wageningen Academic Publishers: Wageningen, The Netherlands, 2020.
- Musaogullari, A.; Chai, Y.C. Redox Regulation by Protein S-Glutathionylation: From Molecular Mechanisms to Implications in Health and Disease. Int. J. Mol. Sci. 2020, 21, 8113.
- Sun, L.; Wang, X.; Saredy, J.; Yuan, Z.; Yang, X.; Wang, H. Innate-adaptive immunity interplay and redox regulation in immune response. Redox. Biol. 2020, 37, 101759.
- Francioso, A.; Baseggio Conrado, A.; Mosca, L.; Fontana, M. Chemistry and Biochemistry of Sulfur Natural Compounds: Key Intermediates of Metabolism and Redox Biology. Oxid. Med. Cell Longev. 2020, 2020, 8294158.
- Corkey, B.E.; Deeney, J.T. The Redox Communication Network as a Regulator of Metabolism. Front. Physiol. 2020, 11, 567796.
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474.
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell. Biol. 2020, 21, 363–383.
- Carvalho, R.H.; Ida, E.I.; Madruga, M.S.; Martínez, S.L.; Shimokomaki, M.; Estévez, M. Underlying connections between the redox system imbalance, protein oxidation and impaired quality traits in pale, soft and exudative (PSE) poultry meat. Food Chem. 2017, 215, 129–137.
- Pan, X.; Zhang, L.; Xing, T.; Li, J.; Gao, F. The impaired redox status and activated Nrf2/ARE pathway in wooden breast myopathy in broiler chickens. Asian-Australas. J. Anim. Sci. 2020, in press.
- Estevez, M.; Petracci, M. Benefits of Magnesium Supplementation to Broiler Subjected to Dietary and Heat Stress: Improved Redox Status, Breast Quality and Decreased Myopathy Incidence. Antioxidants 2019, 8, 456.
- Dalgaard, T.S.; Briens, M.; Engberg, R.M.; Lauridsen, C. The influence of selenium and selenoproteins on immune responses of poultry and pigs. Anim. Feed Sci. Technol. 2018, 238, 73–83.
- Lauridsen, C. From oxidative stress to inflammation: Redox balance and immune system. Poult. Sci. 2019, 98, 4240–4246.
- Surai, P.F. Selenium in Poultry Nutrition and Health; Wageningen Academic Publishers: Wageningen, The Netherlands, 2018.
- Surai, P.F.; Fisinin, V.I. Antioxidant-Prooxidant Balance in the Intestine: Applications in Chick Placement and Pig Weaning. J. Vet. Sci. Med. 2015, 3, 16.
- Mishra, B.; Jha, R. Oxidative Stress in the Poultry Gut: Potential Challenges and Interventions. Front. Vet. Sci. 2019, 6, 60.
- Dong, Y.; Lei, J.; Zhang, B. Effects of dietary quercetin on the antioxidative status and cecal microbiota in broiler chickens fed with oxidized oil. Poult. Sci. 2020, 99, 4892–4903.
- Kövesi, B.; Cserháti, M.; Erdélyi, M.; Zándoki, E.; Mézes, M.; Balogh, K. Long-Term Effects of Ochratoxin A on the Glutathione Redox System and Its Regulation in Chicken. Antioxidants 2019, 8, 178.
- Egresi, A.; Süle, K.; Szentmihályi, K.; Blázovics, A.; Fehér, E.; Hagymási, K.; Fébel, H. Impact of milk thistle (Silybum marianum) on the mycotoxin caused redox-homeostasis imbalance of ducks liver. Toxicon 2020, 187, 181–187.
- Liu, Y.; Zhao, H.; Wang, Y.; Guo, M.; Mu, M.; Xing, M. Arsenic (III) and/or copper (II) induces oxidative stress in chicken brain and subsequent effects on mitochondrial homeostasis and autophagy. J. Inorg. Biochem. 2020, 211, 111201.
- Surai, P.F.; Kochish, I.I. Nutritional modulation of the antioxidant capacities in poultry: The case of selenium. Poult. Sci. 2019, 98, 4231–4239.
- Surai, P.F.; Kochish, I.I.; Romanov, M.N.; Griffin, D.K. Nutritional modulation of the antioxidant capacities in poultry: The case of vitamin E. Poult. Sci. 2019, 98, 4030–4041.
- Surai, P.F.; Kochish, I.I. Carotenoids in Aviculture. In Pigments from Microalgae Handbook; Springer Nature Switzerland: Cham, Switzerland, 2020; pp. 515–540.
- Tolba, S.A.; Magnuson, A.D.; Sun, T.; Lei, X.G. Dietary supplemental microalgal astaxanthin modulates molecular profiles of stress, inflammation, and lipid metabolism in broiler chickens and laying hens under high ambient temperatures. Poult. Sci. 2020, 99, 4853–4860.
- Surai, P.F.; Fisinin, V.I. Vitagenes in poultry production. Part 3. Vitagene concept development. Worlds Poult. Sci. J. 2016, 72, 793–804.
- Surai, P.F.; Fisinin, V.I. Antioxidant system regulation: From vitamins to vitagenes. In Handbook of Cholesterol; Watson, R.R., de Meester, F., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016; pp. 451–481.
- Surai, P.F.; Kochish, I.I.; Fisinin, V.I. Antioxidant systems in poultry biology: Nutritional modulation of vitagenes. Eur. J. Poult. Sci. 2017, 81, 1612–9199.
- Surai, P.F.; Kochish, I.I.; Fisinin, V.I.; Grozina, A.A.; Shatskikh, E.V. Molecular Mechanisms of Chicken Gut Health Maintenance: Role of Microbiota; Agricultural Technologies: Moscow, Russia, 2018.
- Patel, M.; Horgan, P.G.; McMillan, D.C.; Edwards, J. NF-κB pathways in the development and progression of colorectal cancer. Transl. Res. 2018, 197, 43–56.
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 175–190.
- Wang, X.; Hai, C. Novel insights into redox system and the mechanism of redox regulation. Mol. Biol. Rep. 2016, 43, 607–628.
- Dayalan Naidu, S.; Kostov, R.V.; Dinkova-Kostova, A.T. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol. Sci. 2015, 36, 6–14.
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15.
- Cai, D.; Khor, S. “Hypothalamic Microinflammation” Paradigm in Aging and Metabolic Diseases. Cell Metab. 2019, 30, 19–35.
- Yan, J.; Zhang, H.; Yin, Y.; Li, J.; Tang, Y.; Purkayastha, S.; Li, L.; Cai, D. Obesity- and aging-induced excess of central transforming growth factor-beta potentiates diabetic development via an RNA stress response. Nat. Med. 2014, 20, 1001–1008.
- Zhang, X.; Zhang, G.; Zhang, H.; Karin, M.; Bai, H.; Cai, D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell 2008, 135, 61–73.
- Meng, Q.; Cai, D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway. J. Biol. Chem. 2011, 286, 32324–32332.
- Jones, S.V.; Kounatidis, I. Nuclear Factor-Kappa B and Alzheimer Disease, Unifying Genetic and Environmental Risk Factors from Cell to Humans. Front. Immunol. 2017, 8, 1805.
- Kurata, S.; Matsumoto, M.; Tsuji, Y.; Nakajima, H. Lipopolysaccharide activates transcription of the heme oxygenase gene in mouse M1 cells through oxidative activation of nuclear factor kappa B. Eur. J. Biochem. 1996, 239, 566–571.
- Tornatore, L.; Thotakura, A.K.; Bennett, J.; Moretti, M.; Franzoso, G. The nuclear factor kappa B signaling pathway: Integrating metabolism with inflammation. Trends Cell. Biol. 2012, 22, 557–566.
- Mauro, C.; Leow, S.C.; Anso, E.; Rocha, S.; Thotakura, A.K.; Tornatore, L.; Moretti, M.; De Smaele, E.; Beg, A.A.; Tergaonkar, V.; et al. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 2011, 13, 1272–1279.
- Ikeda, T.; Honjo, K.; Hirota, Y.; Onodera, T. Isolation of the chicken NF-kappa B p65 subunit-encoding cDNA and characterization of its products. Gene 1993, 133, 237–242.
- Ikeda, T.; Hirota, Y.; Onodera, T. Isolation of a cDNA encoding the chicken p50B/p97 (Lyt-10) transcription factor. Gene 1994, 138, 193–196.
- Krishnan, V.A.; Schatzle, J.D.; Hinojos, C.M.; Bose, H.R., Jr. Structure and regulation of the gene encoding avian inhibitor of nuclear factor kappa B-alpha. Gene 1995, 166, 261–266.
- Van Phi, L. Transcriptional activation of the chicken lysozyme gene by NF-kappa Bp65 (RelA) and c-Rel, but not by NF-kappa Bp50. Biochem. J. 1996, 313, 39–44.
- Piffat, K.A.; Hrdlicková, R.; Nehyba, J.; Ikeda, T.; Liss, A.; Huang, S.; Sif, S.; Gilmore, T.D.; Bose, H.R., Jr. The chicken RelB transcription factor has transactivation sequences and a tissue-specific expression pattern that are distinct from mammalian RelB. Mol. Cell. Biol. Res. Commun. 2001, 4, 266–275.
- Qiu, Y.; Shen, Y.; Li, X.; Ding, C.; Ma, Z. Molecular cloning and functional characterization of a novel isoform of chicken myeloid differentiation factor 88 (MyD88). Dev. Comp. Immunol. 2008, 32, 1522–1530.
- Zhou, P.; Zeng, Y.; Rao, Z.; Li, Y.; Zheng, H.; Luo, R. Molecular characterization and functional analysis of duck IKKα. Dev. Comp. Immunol. 2021, 115, 103880.
- Kim, Y.; Tian, M. NF-kappaB family of transcription factor facilitates gene conversion in chicken B cells. Mol. Immunol. 2009, 46, 3283–3291.
- Chen, K.; Luo, Z.; Zheng, S.J. Gallus Heat shock cognate protein 70, a novel binding partner of Apoptin. Virol. J. 2011, 8, 324.
- Kogut, M.H.; He, H.; Genovese, K.J. Bacterial toll-like receptor agonists induce sequential NF-κB-mediated leukotriene B4 and prostaglandin E2 production in chicken heterophils. Vet. Immunol. Immunopathol. 2012, 145, 159–170.
- Shinohara, H.; Behar, M.; Inoue, K.; Hiroshima, M.; Yasuda, T.; Nagashima, T.; Kimura, S.; Sanjo, H.; Maeda, S.; Yumoto, N.; et al. Positive feedback within a kinase signaling complex functions as a switch mechanism for NF-κB activation. Science 2014, 344, 760–764.
- Tang, X.; Zhang, L.; Wei, W. Roles of TRAFs in NF-κB signaling pathways mediated by BAFF. Immunol. Lett. 2018, 196, 113–118.
- Yang, H.L.; Feng, Z.Q.; Zeng, S.Q.; Li, S.M.; Zhu, Q.; Liu, Y.P. Molecular cloning and expression analysis of TRAF3 in chicken. Genet. Mol. Res. 2015, 14, 4408–4419.
- Zhai, Y.; Luo, F.; Chen, Y.; Zhou, S.; Li, Z.; Liu, M.; Bi, D.; Jin, H. Molecular characterization and functional analysis of duck TRAF6. Dev. Comp. Immunol. 2015, 49, 1–6.
- Guo, Y.; Xu, Y.; Kang, X.; Meng, C.; Gu, D.; Zhou, Y.; Xiong, D.; Geng, S.; Jiao, X.; Pan, Z. Molecular cloning and functional analysis of TRAF6 from Yangzhou great white goose Anser anser. Dev. Comp. Immunol. 2019, 101, 103435.
- Guo, Y.; Xu, Y.; Xiong, D.; Zhou, Y.; Kang, X.; Meng, C.; Gu, D.; Jiao, X.; Pan, Z. Molecular characterisation, expression and functional feature of TRAF6 in the King pigeon (Columba livia). Innate Immun. 2020, 26, 490–504.
- Zhou, Y.; Kang, X.; Xiong, D.; Zhu, S.; Zheng, H.; Xu, Y.; Guo, Y.; Pan, Z.; Jiao, X. Molecular and functional characterization of pigeon (Columba livia) tumor necrosis factor receptor-associated factor 3. Dev. Comp. Immunol. 2017, 69, 51–59.
- Kang, Y.; Nii, T.; Isobe, N.; Yoshimura, Y. Effects of TLR Ligands on the Expression of Cytokines and Possible Role of NFκB in its Process in the Theca of Chicken Follicles. J. Poult. Sci. 2018, 55, 288–300.
- Chen, S.; Cheng, A.; Wang, M. Innate sensing of viruses by pattern recognition receptors in birds. Vet. Res. 2013, 44, 82.
- Downing, T.; Lloyd, A.T.; O’Farrelly, C.; Bradley, D.G. The differential evolutionary dynamics of avian cytokine and TLR gene classes. J. Immunol. 2010, 184, 6993–7000.
- Gillespie, M.; Shamovsky, V.; D’Eustachio, P. Human and chicken TLR pathways: Manual curation and computer-based orthology analysis. Mamm. Genome 2011, 22, 130–138.
- Farnell, M.B.; Crippen, T.L.; He, H.; Swaggerty, C.L.; Kogut, M.H. Oxidative burst mediated by toll like receptors (TLR) and CD14 on avian heterophils stimulated with bacterial toll agonists. Dev. Comp. Immunol. 2003, 27, 423–429.
- Kannaki, T.R.; Reddy, M.R.; Verma, P.C.; Shanmugam, M. Differential Toll-like receptor (TLR) mRNA expression patterns during chicken embryological development. Anim. Biotechnol. 2015, 26, 130–135.
- Yang, Y.; Jiang, Y.; Yin, Q.; Liang, H.; She, R. Chicken intestine defensins activated murine peripheral blood mononuclear cells through the TLR4-NF-kappaB pathway. Vet. Immunol. Immunopathol. 2010, 133, 59–65.
- Yu, H.; Lu, Y.; Qiao, X.; Wei, L.; Fu, T.; Cai, S.; Wang, C.; Liu, X.; Zhong, S.; Wang, Y. Novel Cathelicidins from Pigeon Highlights Evolutionary Convergence in Avain Cathelicidins and Functions in Modulation of Innate Immunity. Sci. Rep. 2015, 5, 11082.
- Ishige, T.; Hara, H.; Hirano, T.; Kono, T.; Hanzawa, K. Basic characterization of avian NK-lysin (NKL) from the Japanese quail, Coturnix japonica. Anim. Sci. J. 2014, 85, 90–95.
- Kamimura, T.; Isobe, N.; Yoshimura, Y. Effects of inhibitors of transcription factors, nuclear factor-κB and activator protein 1, on the expression of proinflammatory cytokines and chemokines induced by stimulation with Toll-like receptor ligands in hen vaginal cells. Poult. Sci. 2017, 96, 723–730.
- Yong, Y.H.; Liu, S.F.; Hua, G.H.; Jia, R.M.; Gooneratne, R.; Zhao, Y.T.; Liao, M.; Ju, X.H. Goose toll-like receptor 3 (TLR3) mediated IFN-γ and IL-6 in anti-H5N1 avian influenza virus response. Vet. Immunol. Immunopathol. 2018, 197, 31–38.
- Gao, M.; Guo, Y.; Du, J.; Song, Z.; Luo, X.; Wang, J.; Han, W. Evolutional conservation of molecular structure and antiviral function of a type I interferon, IFN-kappa, in poultry. Dev. Comp. Immunol. 2018, 89, 44–53.
- Jia, H.; Li, G.; Li, J.; Tian, Y.; Wang, D.; Shen, J.; Tao, Z.; Xu, J.; Lu, L. Cloning, expression and bioinformatics analysis of the duck TLR 4 gene. Br. Poult. Sci. 2012, 53, 190–197.
- Fang, Q.; Pan, Z.; Geng, S.; Kang, X.; Huang, J.; Sun, X.; Li, Q.; Cai, Y.; Jiao, X. Molecular cloning, characterization and expression of goose Toll-like receptor 5. Mol. Immunol. 2012, 52, 117–124.
- Yong, Y.; Liu, S.; Hua, G.; Jia, R.; Zhao, Y.; Sun, X.; Liao, M.; Ju, X. Identification and functional characterization of Toll-like receptor 2-1 in geese. BMC Vet. Res. 2015, 11, 108.
- Xiong, D.; Song, L.; Pan, Z.; Jiao, X. Molecular cloning, characterization, and functional analysis of pigeon (Columba livia) Toll-like receptor 5. Poult. Sci. 2018, 97, 4031–4039.
- Li, S.; Wang, Y.; Zhao, H.; Shao, Y.; Liu, J.; Xing, M. Characterization, functional and signaling elucidation of pigeon (Columba livia) interferon-α: Knockdown p53 negatively modulates antiviral response. Dev. Comp. Immunol. 2019, 90, 29–40.
- Xiong, D.; Song, L.; Pan, Z.; Chen, X.; Geng, S.; Jiao, X. Identification and immune functional characterization of pigeon TLR7. Int. J. Mol. Sci. 2015, 16, 8364–8381.
- Tao, Z.Y.; Zhu, C.H.; Shi, Z.H.; Song, C.; Xu, W.J.; Song, W.T.; Zou, J.M.; Qin, A.J. Molecular characterization, expression, and functional analysis of NOD1 in Qingyuan partridge chicken. Genet. Mol. Res. 2015, 14, 2691–2701.
- Li, H.; Jin, H.; Li, Y.; Liu, D.; Foda, M.F.; Jiang, Y.; Luo, R. Molecular cloning and functional characterization of duck nucleotide-binding oligomerization domain 1 (NOD1). Dev. Comp. Immunol. 2017, 74, 82–89.
- Truong, A.D.; Hong, Y.; Hoang, C.T.; Lee, J.; Hong, Y.H. Chicken IL-26 regulates immune responses through the JAK/STAT and NF-κB signaling pathways. Dev. Comp. Immunol. 2017, 73, 10–20.
- Truong, A.D.; Hong, Y.; Rengaraj, D.; Lee, J.; Lee, K.; Hong, Y.H. Identification and functional characterization, including cytokine production modulation, of the novel chicken Interleukin-11. Dev. Comp. Immunol. 2018, 87, 51–63.
- Hoang, C.T.; Hong, Y.; Truong, A.D.; Lee, J.; Lee, K.; Hong, Y.H. Molecular cloning of chicken interleukin-17B, which induces proinflammatory cytokines through activation of the NF-κB signaling pathway. Dev. Comp. Immunol. 2017, 74, 40–48.
- Rohde, F.; Schusser, B.; Hron, T.; Farkašová, H.; Plachý, J.; Härtle, S.; Hejnar, J.; Elleder, D.; Kaspers, B. Characterization of Chicken Tumor Necrosis Factor-α, a Long Missed Cytokine in Birds. Front. Immunol. 2018, 9, 605.
- Ji, G.G.; Shu, J.T.; Zhang, M.; Ju, X.J.; Shan, Y.J.; Liu, Y.F.; Tu, Y.J. Transcriptional regulatory region and DNA methylation analysis of TNNI1 gene promoters in Gaoyou duck skeletal muscle (Anas platyrhynchos domestica). Br. Poult. Sci. 2019, 60, 202–208.
- Barjesteh, N.; Taha-Abdelaziz, K.; Kulkarni, R.R.; Sharif, S. Innate antiviral responses are induced by TLR3 and TLR4 ligands in chicken tracheal epithelial cells: Communication between epithelial cells and macrophages. Virology 2019, 534, 132–142.
- Sutton, K.M.; Hu, T.; Wu, Z.; Siklodi, B.; Vervelde, L.; Kaiser, P. The functions of the avian receptor activator of NF-κB ligand (RANKL) and its receptors, RANK and osteoprotegerin, are evolutionarily conserved. Dev. Comp. Immunol. 2015, 51, 170–184.
- Truong, A.D.; Hong, Y.; Nguyen, H.T.; Nguyen, C.T.; Chu, N.T.; Tran, H.T.T.; Dang, H.V.; Lillehoj, H.S.; Hong, Y.H. Molecular identification and characterisation of a novel chicken leukocyte immunoglobulin-like receptor A5. Br. Poult. Sci. 2020, in press.




