
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mariko Ishibashi | + 2975 word(s) | 2975 | 2021-01-21 09:00:29 | | | |
| 2 | Vivi Li | + 3 word(s) | 2978 | 2021-01-26 08:39:45 | | |
Video Upload Options
The signaling lymphocytic activation molecule (SLAM) family receptors are expressed on various immune cells and malignant plasma cells in multiple myeloma (MM) patients. In immune cells, most SLAM family molecules bind to themselves to transmit co-stimulatory signals through the recruiting adaptor proteins SLAM-associated protein (SAP) or Ewing’s sarcoma-associated transcript 2 (EAT-2), which target immunoreceptor tyrosine-based switch motifs in the cytoplasmic regions of the receptors. Notably, SLAMF2, SLAMF3, SLAMF6, and SLAMF7 are strongly and constitutively expressed on MM cells that do not express the adaptor proteins SAP and EAT-2.
1. Introduction
Multiple myeloma (MM) is a B-cell malignancy that arises from the clonal expansion of aberrant plasma cells (MM cells). In addition to the accumulation of clonal plasma cells, MM is characterized by elevated levels of monoclonal protein (M-protein), renal failure, hypercalcemia, anemia, and bone lesions [1][2]. This malignancy is caused by multiple genetic alterations and microenvironmental changes in bone marrow (BM) cells and nearly always develops from monoclonal gammopathy of undetermined significance (MGUS), a precancerous condition [1][3][4][5].
In the past several years, the treatment options for patients with refractory MM have advanced dramatically due to the emergence and approval of pharmaceuticals such as immunomodulatory drugs (IMiDs; e.g., lenalidomide and pomalidomide), proteasome inhibitors (e.g., bortezomib, carfilzomib, and ixazomib), and monoclonal antibodies (e.g., elotuzumab, daratumumab, and isatuximab) [1][6][7][8][9][10]. Many clinical trials of immunotherapies for MM are ongoing, with a particular focus on IMiDs, monoclonal antibodies, bispecific antibodies, immune checkpoint inhibitors, and chimeric antigen receptor (CAR) T-cell therapy. Several molecules have been selected as candidate targets for MM immunotherapies, including signaling lymphocytic activation molecule (SLAM) family 7 (SLAMF7) and CD38, which are targeted by elotuzumab and daratumumab/isatuximab, respectively, and other cell-surface antigens such as B-cell maturation antigen (BCMA), CD56, CD138, CD74, interleukin (IL)-6 receptor, vascular endothelial growth factor (VEGF), and activated conformation of integrin β7 [11][12][13].
Promising therapeutic target antigens for MM must have the following characteristics: (1) specific or strong expression on the surfaces of MM cells relative to normal immune cells and tissues; (2) expression on MM progenitor cells or cancer stem cells but not on normal hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs); (3) constitutive strong expression, regardless of disease progression or treatment; and (4) an association with the biological functions or molecular pathogenesis of MM (e.g., cell survival and growth, antiapoptotic responses, angiogenesis, cell–cell communications, and interactions with BM stromal cells). Therefore, an understanding of the expression and biological functions of these molecules in MM is important to support the development of novel therapeutic targets and the prevention of treatment-related side effects.
2. Overview of SLAM Family Receptors
SLAM family receptors play important roles in the immune responses mediated by a broad range of innate and adaptive immune cells [14][15][16][17][18]. The SLAM family comprises nine members: SLAMF1 (CD150); SLAMF2 (CD48); SLAMF3 (CD229, Ly9); SLAMF4 (CD244, 2B4); SLAMF5 (CD84); SLAMF6 (CD352, NTB-A, SF2000); SLAMF7 (CD319, CS1, CRACC); SLAMF8 (CD353); and SLAMF9 (CD84H1, SF2001) (Figure 1). All except for SLAMF2 are type I transmembrane glycoproteins within the immunoglobulin (Ig) superfamily and are composed of extracellular IgV- and IgC2-like domains with high self-affinity through the IgV-like domain. In contrast, SLAMF2 interacts with SLAMF4 [14][19][20]. The cytoplasmic tail of a SLAM family receptor contains an immunoreceptor tyrosine-based switch motif (ITSM), TxYxxI/V, where T, Y, I, V, and x represent threonine, tyrosine, isoleucine, valine, and any other amino acid, respectively (Figure 1) [19][20]. Self-ligated SLAM family receptors recruit Src homology region 2 (SH2) domain-containing adaptor proteins such as SLAM-associated protein (SAP; SH2D1A) or Ewing’s sarcoma-associated transcript 2 (EAT-2; SH2D1B) to the phosphorylated tyrosine within the ITSM sequence. SAP is expressed in T cells, natural killer (NK) cells, NKT cells, eosinophils, and platelets but is absent or minimally expressed in some B cell populations, macrophages, and dendritic cells (DCs) [16][21]. EAT-2 is expressed in NK cells and antigen-presenting cells (APCs) including B cells, macrophages, and DCs [22][23]. In SLAM family receptors containing ITSMs in the cytoplasmic domain (SLAMF1 and SLAMF3–SLAMF7), SLAM–SAP, and SLAM–EAT-2 complexes interact directly with Src family kinase Fyn and phospholipase Cγ (PLCγ), respectively, and trigger an activating signal within these immune cells (Figure 1). However, the signal transduction pathway mediated by SLAMF2, SLAMF8, and SLAMF9, which lack intercellular signaling motifs, is still unknown. Further, SAP and EAT-2 inhibit the recruitment of inhibitory adaptor proteins such as SH2 domain phosphatase-1 (SHP-1), SHP-2, SH-2 domain inositol 5ʹ-polyphosphatase 1 (SHIP1), or Csk to ITSMs of SLAM family receptors and, in the absence of SAP and EAT-2, the self-ligated receptor transduces an inhibitory signal via these inhibitory adaptor proteins [24][25][26].
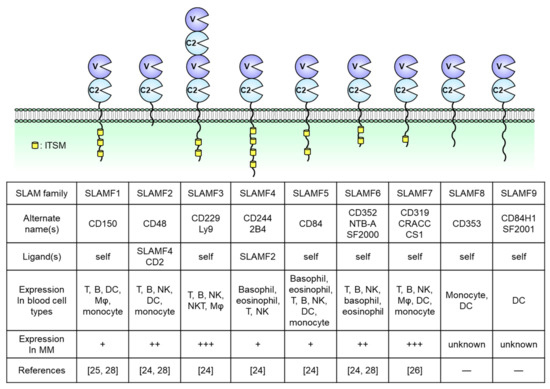
Figure 1. Structures of nine signaling lymphocytic activation molecule (SLAM) family receptors. The extracellular regions contain IgV (V)- and IgC2 (C2)-like domains. The receptors bind to self-ligands with high affinity through the IgV-like domain. In all family members except for SLAMF2, SLAMF8, and SLAMF9, the cytoplasmic tail contains an immunoreceptor tyrosine-based switch motif (ITSM). The expression of immune cells is from the database of the Human Protein Atlas (https://www.proteinatlas.org/). DC, dendritic cell, Mφ, macrophage, NK, natural killer.
SLAM family molecules are expressed in various hematological malignancies (e.g., MM, chronic lymphocytic leukemia [CML], acute myeloid leukemia [AML], lymphoma) and solid tumors (e.g., hepatocellular carcinoma, central nervous system tumors, renal cancer) [19]. In MM patients, malignant plasma cells strongly express SLAMF2, SLAMF3, SLAMF6, and SLAMF7. In contrast, SLAMF1, SLAMF4, and SLAMF5 expression is limited and SLAMF8 and SLAMF9 have not been reported in this malignancy [27][28][29][30][31]. The genes encoding SLAM family receptors and EAT-2 are clustered in the long arm of chromosome 1 (1q23) [17][32]. In contrast, the gene encoding SAP is localized on the X chromosome (Xq25), and X-linked lymphoproliferative disease (XLP), a rare heritable immunodeficiency disorder, is associated with SAP deficiency [32][33]. Although 36% of newly diagnosed MM patients present with a gain or amplification of the long arm of chromosome 1 (+1q; region, 1q21–1q23.1) [34][35][36], the relationship between +1q and the level of SLAM family receptor expression has not been explored previously. This review focuses on the expression and biological functions of SLAMF2, SLAMF3, SLAMF6, and SLAMF7 in both normal immune cells and MM cells and discusses their potential usefulness as therapeutic targets in relapsed/refractory MM.
3. SLAMF2 (CD48)
SLAMF2 is a glycosylphosphatidylinositol (GPI)-anchored glycan of the CD2 superfamily of Ig-like receptors and is expressed on nearly all hematopoietic cells [37]. Unlike other SLAM family molecules, SLAMF2 facilitates cell–cell communication by binding to its ligands CD2 and SLAMF4 (also known as CD244 and 2B4) on other immune cells, such as T, B, and NK cells [37][38][39]. The binding affinity of human SLAMF2 for SLAMF4 is 10-fold higher than its affinity for CD2 [39]. In T cells, SLAMF2 binds to GPI in cell membrane lipid rafts, where it activates leukocyte C-terminal Src kinase (Lck) to promote T cell receptor (TCR) signaling and activation (Figure 2A) [40]. When expressed on APCs such as DCs, SLAMF2 serves as a strong co-stimulatory molecule to enhance the cytotoxicity of SLAMF4-expressing effector/memory CD8+ T cells, and this cell–cell interaction prolongs DC survival [37][41]. Furthermore, SLAMF2 expressed on DCs activates SLAMF4-expressing NK cells via PLCγ activation, Ca2+ flux induction, and SAP- or EAT-2-mediated ERK activation.
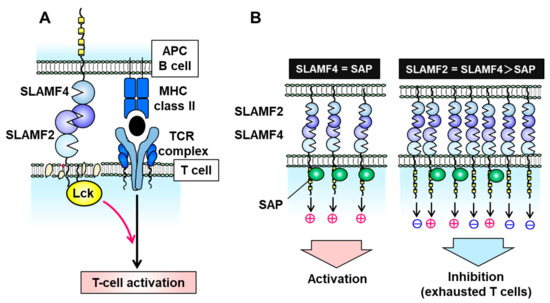
Figure 2. Signal transduction via the signaling lymphocytic activation molecule family receptor (SLAMF) 2–SLAMF4 interaction. (A) On T cells, SLAMF2 promotes T cell receptor (TCR) signaling and T cell activation by binding to SLAMF4 on antigen-presenting cells (APCs) and activating the tyrosine kinase Lck in T cell membrane lipid rafts. (B) The SLAMF2–SLAMF4 interaction facilitates natural killer (NK) or T cell activation in the presence of sufficient levels of the adaptor protein SAP but induces an inhibitory signal in the absence of SAP.
In the absence of SAP and EAT-2, the SLAMF2–SLAMF4 interaction induces an inhibitory signal in NK cells via SHP1, SHP2, and SHIP-1 [17][26][42][43][44]. SLAMF2 and SLAMF4 on immune cells are upregulated under inflammatory conditions of the tumor microenvironment and chronic viral infection [37][44]. SLAMF4-overexpressing T cells exhibit a phenotype of exhausted CD8+ T cells which expresses immune checkpoint inhibitory receptors such as PD-1, LAG-3, CD160, CTLA-4, and TIM-3 and decreases IL-2 and interferon (IFN)-γ production [44]. Inflammatory conditions can induce an imbalance in the expression of SLAMF4 and SAP (e.g., high SLAMF4 and normal SAP) which suppresses T cell activation and induces T cell exhaustion (Figure 2B). Exhausted T and NK cells have been observed in various malignancies, including MM, melanoma, hepatocellular carcinoma, and acute myelocytic leukemia (AML) [45][46][47][48]. Zelle-Rieser et al. showed that SLAMF4-positive exhausted T cells exhibited characteristics such as a lower proliferative capacity and impaired function in the MM microenvironment [45]. Therefore, the SLAMF2–SLAMF4 interaction has both co-stimulatory and co-inhibitory functions in immune cells, depending on the status of SAP or EAT-2 expression (Figure 2B).
Hosen et al. reported that SLAMF2 was strongly expressed on cancer cells from 22 of 24 patients with MM but not on normal HSCs and HPCs from those individuals [49]. In that study, an antihuman CD48 antibody exerted cytotoxic effects against MM cells via NK cell-mediated antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) but did not target CD34+ HSCs/HPCs [49]. The antibody eliminated human myeloma cell lines implanted under the skin of immunodeficient mice [49]. Furthermore, in a study of 79 patients with MM, Ashour et al. observed the maintenance of high SLAMF2 expression levels when comparing cases in the following categories: newly diagnosed versus relapsed/refractory MM; Revised International Staging System (R-ISS) stage I/II versus stage III; with versus without intensive treatment; and responsiveness versus nonresponsiveness to bortezomib or IMiDs [50]. Given this broad expression of SLAMF2 in most MM patients, Lewis and colleagues evaluated the cytotoxic effect of SGN-CD48A, a CD48-targeting antibody–drug conjugate (ADC) with a glucuronide–monomethylauristatin E (MMAE) linker [51]. Exposure to SGN-CD48A potently induced G2/M cell cycle arrest in MM cells in vitro and led to complete remission in nearly all tested U266 or NCI-H929 xenograft-bearing mice but had minimal effects on human B, T, and NK cells [51]. Interestingly, however, SGN-CD48A killed both CD48+ and CD48KO MM cells in an admixed co-culture, suggesting a potential cytotoxic bystander effect mediated by the activation of immune cell death [52]. The combination of SGN-CD48A and daratumumab yielded increased antitumor activity relative to SGN-CD48 alone or in combination with elotuzumab in MM xenograft mouse models [52]. Those results suggest that SGN-CD48A may exert cytotoxicity against MM cells through multiple direct and indirect mechanisms and may be a candidate for combination therapy with daratumumab. A clinical study of SGN–CD48A monotherapy for relapsed/refractory MM patients is in progress after patient recruitment ended (NCT02954796). Although these SLAMF2-targeting therapies induced significant effects in preclinical studies, it will be necessary to clarify whether the expression of SLAMF2 on MM cells is associated with the pathogenesis of this malignancy.
4. SLAMF3 (CD229, Ly9)
SLAMF3 is expressed strongly on most MM cell lines, and Atanackovic et al. identified this receptor as the most intensely expressed phosphorylated immunoreceptor in a human phosphoimmunoreceptor array involving the MM cell line MOLP-8 [27]. SLAMF3 is also expressed on Sca-1+c-kit+Lin− HSCs and mainly localizes to T and B lymphocytes and NK and NKT cells (Figure 1) [53][54][55]. In contrast to the other SLAM family receptors, SLAMF3 contains a unique extracellular domain that comprises two tandem repeats of IgV–IgC2 and interacts homologically with itself via the N-terminal IgV domain (Figure 3A) [54][56]. The cytoplasmic domain of SLAMF3 contains two ITSMs. In T cells, SLAMF3 recruits SAP through its SH2 domain, and SAP then interacts with the Src family kinase Fyn to regulate protein kinase Cθ (PKCθ)–NF-κB pathway activity (Figure 3A) [55][57][58][59]. In the presence of an anti-mouse SLAMF3 antibody (Ly9.ab3), the inability of SLAMF3 to self-ligate suppresses the expression of the activation markers CD25 and CD69 on anti-CD3/anti-CD28 antibody-stimulated murine T cells and further decreases the secretion of IFN-γ, IL-2, IL-4, IL-6, IL-10, and tumor necrosis factor (TNF)-α [53]. SLAMF3 is associated with enhanced T cell activation and helper T cell type 2 (Th2) polarization, and consequently SLAMF3 deficiency markedly reduces T cell proliferation and IL-2 production and causes a mild Th2 defect [60]. Thus SLAMF3 is associated with enhanced T cell activation and promotes Th2 polarization.
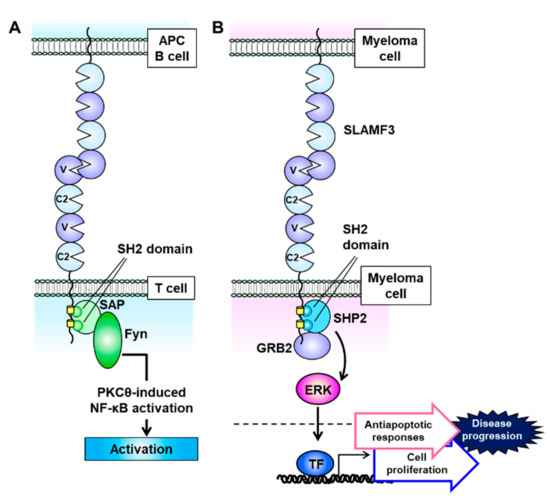
Figure 3. Signaling lymphocytic activation molecule family receptor (SLAMF) 3 signal transduction is mediated via self-ligation and cell–cell interactions. (A) SLAMF3 recruits SAP at two immunoreceptor tyrosine-based switch motif (ITSM) domains, and the SLAM–SAP–Fyn signaling pathway promotes T cell activation. (B) On multiple myeloma (MM) cells, SLAMF3 activates the ERK signal transduction pathway. This function is mediated via self-ligand interactions between MM cells and SHP2 and GRB2 formation, which accelerates cell proliferation and antiapoptotic signaling to increase the malignant potential of MM.
SLAMF3 is expressed at higher levels on malignant plasma cells from MM patients than on other BM hematopoietic cells in the same samples [27][61][62][63]. However, SLAMF3 is also strongly expressed on normal plasma cells from MM patients and healthy controls [27][61][62][63][64]. Yousef et al. demonstrated that SLAMF3 expression levels were unaltered in 17 patients with MGUS, 49 with MM, 7 with smoldering MM (SMM), and 4 with plasma cell leukemia [62]. Similarly, we observed that the expression of SLAMF3 remained constitutively strong, regardless of disease stage, in a study of 17 patients with MGUS, 153 with newly diagnosed MM (19 asymptomatic and 134 symptomatic), and 30 with relapsed/refractory MM. In contrast, there were significant decreases in the expression of the plasma cell-surface antigen CD138 with disease progression [63]. Atanackovic et al. used small interfering RNA (siRNA) to demonstrate that SLAMF3 knockdown reduced the proliferation and survival of MM cell lines and enhanced apoptosis induced by anti-MM agents [27]. Similarly, we confirmed that SLAMF3 knockdown and knockout, which were, respectively, achieved using small hairpin RNA (shRNA) and the CRISPR/Cas9 system, nearly abolished the aggressive phenotypic characteristics of MM, while SLAMF3 overexpression promoted those characteristics [63].
Notably, MM cell lines do not express SAP and EAT-2, and therefore SLAMF3 signaling pathway activity is closely related to the recruitment of the adaptor proteins SHP2 and growth factor receptor-bound protein 2 (GRB2), which are expressed in most cell lines [63]. SHP2 and GRB2, respectively, bind to the two ITSMs and a C-terminal end motif (amino acid sequence: YENE) in the cytoplasmic domain of SLAMF3 and interact with each other (Figure 3B) [63]. When expressed on MM cells, SLAMF3 transmits MAPK/ERK signals mediated by the recruitment of SHP2 and GRB2 via self-ligand interactions and subsequently promotes antiapoptotic responses and cell proliferation (Figure 3B) [63]. The GRB2-mediated signaling cascade promotes cell proliferation and survival in multiple types of malignancy [65]. GRB2 is expressed much more strongly in plasma cells from MM patients relative to healthy controls, despite nearly equal SLAMF3 expression levels [63]. These results suggest that the SLAMF3–SHP2–GRB2 signaling pathway strongly promotes the aggressive MM cell phenotype.
The gene encoding SLAMF3 contains a single nucleotide polymorphism (SNP), rs509749 (1804A>G, M602V), which causes a nonsynonymous change from M (codon ATG) to V (codon CTG) at amino acid residue 602 (M602V) within ITSM1 of the intracellular domain. The frequencies of the rs509749 A and G alleles are 0.34 and 0.66, respectively, in the global population. The G allele is the major type in all but European populations, in which the frequencies are nearly equal (A = 0.55, G = 0.45) [66]. An association between the A allele and susceptibility to systemic lupus erythematosus (SLE) has been reported in British and Canadian populations [67][68]. We studied 68 Japanese patients with MM to determine the rs509749 allelic frequency in this population. The G allele was dominant (G = 0.76), and the frequencies of the GG, GA, and AA genotypes were 61.8%, 29.4%, and 8.8%, respectively, which were very similar to those in healthy Japanese controls [69]. Interestingly, MM patients harboring the rs509749 GG genotype had significantly shorter overall survival durations than those harboring the GA and AA genotypes (p = 0.046). We further determined that in MM cells, the A allele of rs509749 was associated with a markedly lower SLAMF3 binding affinity for SHP2 and GRB2 when compared with the G allele, and this reduced affinity led to decreases in ERK activation and aggressive MM behaviors [69]. Therefore, the G allele of the SLAMF3 SNP rs509749 might be a risk factor for MM development but not for plasma cell tumorigenesis.
We further investigated whether SLAMF3 may be a promising serum biomarker in MM patients. Notably, we observed elevated serum concentrations of soluble SLAMF3 (sSLAMF3) with MM progression, which led to markedly high levels in patients with advanced-stage disease [63]. sSLAMF3 is produced by cleavage of the extracellular domain from SLAMF3-expressing MM cells via matrix metalloproteinase-9 (MMP-9) or other enzymatic mechanisms [63]. MM patients harboring high circulating levels of sSLAMF3 exhibit more aggressive clinical characteristics and a significantly shorter progression-free survival (PFS) duration than those with low levels. These results suggest that the serum level of sSLAMF3 might reflect the progression of MM and could thus be a useful new prognostic marker.
SLAMF3 is expressed on MM cells regardless of disease progression or cell phenotype and is detected not only on CD38+CD138+ plasma cells but also on aberrant CD56+ plasma cells and CD38+CD138low/negative chemotherapy-resistant myeloma progenitor cells [62][63]. Therefore, SLAMF3 might be useful as both a cell-surface marker and an immunotherapeutic target in MM. A monoclonal anti-SLAMF3 antibody was shown to induce MM cell lysis effectively via ADCC and CDC [27]. Moreover, the blockade of SLAMF3-mediated self-ligation between MM cells suppresses cell proliferation and facilitates melphalan-induced apoptosis [63]. Atanackovic’s group developed an innovative CAR-T cell therapy specific for SLAMF3 on MM cells [70]. Those CAR-T cells efficiently killed SLAMF3high plasma cells but not SLAMF3low T cells or native B cells. Interestingly, SLAMF3 CAR-T cells more effectively eliminated SLAMF3-positive, BCMA-negative memory B cells (which include putative clonotypic MM progenitor cells) when compared with BCMA CAR-T cells [70]. Thus, SLAMF3-targeted CAR-T therapy could potentially prevent posttreatment MM relapse, unlike BCMA CAR-T therapy. SLAMF3-targeted strategies have not yet proceeded to preclinical trials, and the potential for strong and promising effects in MM patients remains to be demonstrated.
References
- Rajkumar, S.V. Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018, 93.
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060.
- Prideaux, S.M.; Conway-O’Brien, E.; Chevassut, T.J. The genetic architecture of multiple myeloma. Adv. Hematol. 2014, 2014, 864058.
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046.
- Gooding, S.; Edwards, C.M. New approaches to targeting the bone marrow microenvironment in multiple myeloma. Curr. Opin. Pharmacol. 2016, 28, 43–49.
- Tamura, H.; Ishibashi, M.; Sunakawa, M.; Inokuchi, K. Immunotherapy for Multiple Myeloma. Cancers 2019, 11, 9.
- Swan, D.; Lynch, K.; Gurney, M.; O’Dwyer, M. Current and emerging immunotherapeutic approaches to the treatment of multiple myeloma. Ther. Adv. Hematol. 2019, 10.
- Moreau, P.; Zamagni, E.; Mateos, M.V. Treatment of patients with multiple myeloma progressing on frontline-therapy with lenalidomide. Blood Cancer J. 2019, 9, 38.
- Neri, P.; Bahlis, N.J.; Lonial, S. New Strategies in Multiple Myeloma: Immunotherapy as a Novel Approach to Treat Patients with Multiple Myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5959–5965.
- Anderson, K.C. Progress and Paradigms in Multiple Myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5419–5427.
- Ma, J.; Li, Q.; Yu, Z.; Cao, Z.; Liu, S.; Chen, L.; Li, H.; Gao, S.; Yan, T.; Wang, Y.; et al. Immunotherapy Strategies Against Multiple Myeloma. Technol. Cancer Res. Treat. 2017, 16, 717–726.
- Hosen, N.; Matsunaga, Y.; Hasegawa, K.; Matsuno, H.; Nakamura, Y.; Makita, M.; Watanabe, K.; Yoshida, M.; Satoh, K.; Morimoto, S.; et al. The activated conformation of integrin β(7) is a novel multiple myeloma-specific target for CAR T cell therapy. Nat. Med. 2017, 23, 1436–1443.
- Shah, N.; Chari, A.; Scott, E.; Mezzi, K.; Usmani, S.Z. B-cell maturation antigen (BCMA) in multiple myeloma: Rationale for targeting and current therapeutic approaches. Leukemia 2020, 34, 985–1005.
- Veillette, A.; Latour, S. The SLAM family of immune-cell receptors. Curr. Opin. Immunol. 2003, 15, 277–285.
- Veillette, A. SLAM-family receptors: Immune regulators with or without SAP-family adaptors. Cold Spring Harb. Perspect. Biol. 2010, 2, a002469.
- Schwartzberg, P.L.; Mueller, K.L.; Qi, H.; Cannons, J.L. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 2009, 9, 39–46.
- Cannons, J.L.; Tangye, S.G.; Schwartzberg, P.L. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 2011, 29, 665–705.
- Ma, C.S.; Deenick, E.K. The role of SAP and SLAM family molecules in the humoral immune response. Ann. N. Y. Acad. Sci. 2011, 1217, 32–44.
- Fouquet, G.; Marcq, I.; Debuysscher, V.; Bayry, J.; Rabbind-Singh, A.; Bengrine, A.; Nguyen-Khac, E.; Naassila, M.; Bouhlal, H. Signaling lymphocytic activation molecules Slam and cancers: Friends or foes? Oncotarget 2018, 9, 16248–16262.
- Detre, C.; Keszei, M.; Romero, X.; Tsokos, G.C.; Terhorst, C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin. Immunopathol. 2010, 32, 157–171.
- Wu, N.; Veillette, A. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 2016, 38, 45–51.
- Morra, M.; Lu, J.; Poy, F.; Martin, M.; Sayos, J.; Calpe, S.; Gullo, C.; Howie, D.; Rietdijk, S.; Thompson, A.; et al. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J. 2001, 20, 5840–5852.
- Pérez-Quintero, L.A.; Roncagalli, R.; Guo, H.; Latour, S.; Davidson, D.; Veillette, A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J. Exp. Med. 2014, 211, 727–742.
- Dong, Z.; Veillette, A. How do SAP family deficiencies compromise immunity? Trends Immunol. 2010, 31, 295–302.
- Le Borgne, M.; Shaw, A.S. SAP signaling: A dual mechanism of action. Immunity 2012, 36, 899–901.
- Chen, S.; Dong, Z. NK cell recognition of hematopoietic cells by SLAM-SAP families. Cell. Mol. Immunol. 2019, 16, 452–459.
- Atanackovic, D.; Panse, J.; Hildebrandt, Y.; Jadczak, A.; Kobold, S.; Cao, Y.; Templin, J.; Meyer, S.; Reinhard, H.; Bartels, K.; et al. Surface molecule CD229 as a novel target for the diagnosis and treatment of multiple myeloma. Haematologica 2011, 96, 1512–1520.
- Gordiienko, I.M.; Shlapatska, L.M.; Kovalevska, L.M.; Sidorenko, S.P. Differential expression of CD150/SLAMF1 in normal and malignant B cells on the different stages of maturation. Exp. Oncol. 2016, 38, 101–107.
- Hsi, E.D.; Steinle, R.; Balasa, B.; Szmania, S.; Draksharapu, A.; Shum, B.P.; Huseni, M.; Powers, D.; Nanisetti, A.; Zhang, Y.; et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 2775–2784.
- Radhakrishnan, S.V.; Bhardwaj, N.; Luetkens, T.; Atanackovic, D. Novel anti-myeloma immunotherapies targeting the SLAM family of receptors. Oncoimmunology 2017, 6, e1308618.
- Muccio, V.E.; Saraci, E.; Gilestro, M.; Gattei, V.; Zucchetto, A.; Astolfi, M.; Ruggeri, M.; Marzanati, E.; Passera, R.; Palumbo, A.; et al. Multiple myeloma: New surface antigens for the characterization of plasma cells in the era of novel agents. Cytom. Part B Clin. Cytom. 2016, 90, 81–90.
- Engel, P.; Eck, M.J.; Terhorst, C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat. Rev. Immunol. 2003, 3, 813–821.
- Sayos, J.; Wu, C.; Morra, M.; Wang, N.; Zhang, X.; Allen, D.; van Schaik, S.; Notarangelo, L.; Geha, R.; Roncarolo, M.G.; et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 1998, 395, 462–469.
- Walker, B.A.; Leone, P.E.; Chiecchio, L.; Dickens, N.J.; Jenner, M.W.; Boyd, K.D.; Johnson, D.C.; Gonzalez, D.; Dagrada, G.P.; Protheroe, R.K.; et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010, 116, e56–e65.
- Smetana, J.; Oppelt, J.; Štork, M.; Pour, L.; Kuglík, P. Chromothripsis 18 in multiple myeloma patient with rapid extramedullary relapse. Mol. Cytogenet. 2018, 11, 7.
- Sklavenitis-Pistofidis, R.; Reidy, M.; Huynh, D.; Salem, K.Z.; Park, J.; Glavey, S.; Leleu, X.; Roo, D.; Ghobrial, I.M.; Manier, S. Founding Precision Therapy in 1q-Amplified Multiple Myeloma. Blood 2018, 132 (Suppl. 1), 1007.
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20.
- Brown, M.H.; Boles, K.; van der Merwe, P.A.; Kumar, V.; Mathew, P.A.; Barclay, A.N. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998, 188, 2083–2090.
- Arulanandam, A.R.; Moingeon, P.; Concino, M.F.; Recny, M.A.; Kato, K.; Yagita, H.; Koyasu, S.; Reinherz, E.L. A soluble multimeric recombinant CD2 protein identifies CD48 as a low affinity ligand for human CD2: Divergence of CD2 ligands during the evolution of humans and mice. J. Exp. Med. 1993, 177, 1439–1450.
- Moran, M.; Miceli, M.C. Engagement of GPI-linked CD48 contributes to TCR signals and cytoskeletal reorganization: A role for lipid rafts in T cell activation. Immunity 1998, 9, 787–796.
- Kis-Toth, K.; Tsokos, G.C. Engagement of SLAMF2/CD48 prolongs the time frame of effective T cell activation by supporting mature dendritic cell survival. J. Immunol. 2014, 192, 4436–4442.
- Meinke, S.; Watzl, C. NK cell cytotoxicity mediated by 2B4 and NTB-A is dependent on SAP acting downstream of receptor phosphorylation. Front. Immunol. 2013, 4, 3.
- Morandi, B.; Costa, R.; Falco, M.; Parolini, S.; de Maria, A.; Ratto, G.; Mingari, M.C.; Melioli, G.; Moretta, A.; Ferlazzo, G. Distinctive lack of CD48 expression in subsets of human dendritic cells tunes NK cell activation. J. Immunol. 2005, 175, 3690–3697.
- Agresta, L.; Hoebe, K.H.N.; Janssen, E.M. The Emerging Role of CD244 Signaling in Immune Cells of the Tumor Microenvironment. Front. Immunol. 2018, 9, 2809.
- Zelle-Rieser, C.; Thangavadivel, S.; Biedermann, R.; Brunner, A.; Stoitzner, P.; Willenbacher, E.; Greil, R.; Jöhrer, K. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J. Hematol. Oncol. 2016, 9, 116.
- Baitsch, L.; Baumgaertner, P.; Devêvre, E.; Raghav, S.K.; Legat, A.; Barba, L.; Wieckowski, S.; Bouzourene, H.; Deplancke, B.; Romero, P.; et al. Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients. J. Clin. Investig. 2011, 121, 2350–2360.
- Wu, Y.; Kuang, D.M.; Pan, W.D.; Wan, Y.L.; Lao, X.M.; Wang, D.; Li, X.F.; Zheng, L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology 2013, 57, 1107–1116.
- Tan, J.; Chen, S.; Lu, Y.; Yao, D.; Xu, L.; Zhang, Y.; Yang, L.; Chen, J.; Lai, J.; Yu, Z.; et al. Higher PD-1 expression concurrent with exhausted CD8+ T cells in patients with de novo acute myeloid leukemia. Chin. J. Cancer Res. = Chung-Kuo Yen Cheng Yen Chiu 2017, 29, 463–470.
- Hosen, N.; Ichihara, H.; Mugitani, A.; Aoyama, Y.; Fukuda, Y.; Kishida, S.; Matsuoka, Y.; Nakajima, H.; Kawakami, M.; Yamagami, T.; et al. CD48 as a novel molecular target for antibody therapy in multiple myeloma. Br. J. Haematol. 2012, 156, 213–224.
- Ashour, R.; Ri, M.; Aly, S.S.; Yoshida, T.; Tachita, T.; Kanamori, T.; Aoki, S.; Kinoshita, S.; Narita, T.; Totani, H.; et al. Expression analysis of two SLAM family receptors, SLAMF2 and SLAMF7, in patients with multiple myeloma. Int. J. Hematol. 2019, 110, 69–76.
- Lewis, T.S.; Olson, D.; Gordon, K.; Sandall, S.; Quick, M.; Finn, M.; Westendorf, L.; Linares, G.; Leiske, C.; Nesterova, A.; et al. SGN-CD48A: A Novel Humanized Anti-CD48 Antibody-Drug Conjugate for the Treatment of Multiple Myeloma. Blood 2016, 76, 4470.
- Olson, D.J.; Liu, B.A.; Zaval, M.; Cao, A.; Gurgel, J.; Cochran, J.; Stevens, N.; Anderson, M.; Lewis, T.S. Additional mechanisms of action of SGN-CD48A in multiple myeloma and improved antitumor activity in combination with daratumumab. Cancer Res. 2018, 78, 5619.
- Sintes, J.; Vidal-Laliena, M.; Romero, X.; Tovar, V.; Engel, P. Characterization of mouse CD229 (Ly9), a leukocyte cell surface molecule of the CD150 (SLAM) family. Tissue Antigens 2007, 70, 355–362.
- Sandrin, M.S.; Henning, M.M.; Lo, M.F.; Baker, E.; Sutherland, G.R.; McKenzie, I.F. Isolation and characterization of cDNA clones for Humly9: The human homologue of mouse Ly9. Immunogenetics 1996, 43, 13–19.
- de la Fuente, M.A.; Tovar, V.; Villamor, N.; Zapater, N.; Pizcueta, P.; Campo, E.; Bosch, J.; Engel, P. Molecular characterization and expression of a novel human leukocyte cell-surface marker homologous to mouse Ly-9. Blood 2001, 97, 3513–3520.
- Romero, X.; Zapater, N.; Calvo, M.; Kalko, S.G.; de la Fuente, M.A.; Tovar, V.; Ockeloen, C.; Pizcueta, P.; Engel, P. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J. Immunol. 2005, 174, 7033–7042.
- Cannons, J.L.; Yu, L.J.; Hill, B.; Mijares, L.A.; Dombroski, D.; Nichols, K.E.; Antonellis, A.; Koretzky, G.A.; Gardner, K.; Schwartzberg, P.L. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity 2004, 21, 693–706.
- Sayós, J.; Martín, M.; Chen, A.; Simarro, M.; Howie, D.; Morra, M.; Engel, P.; Terhorst, C. Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood 2001, 97, 3867–3874.
- Simarro, M.; Lanyi, A.; Howie, D.; Poy, F.; Bruggeman, J.; Choi, M.; Sumegi, J.; Eck, M.J.; Terhorst, C. SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. Int. Immunol. 2004, 16, 727–736.
- Graham, D.B.; Bell, M.P.; McCausland, M.M.; Huntoon, C.J.; van Deursen, J.; Faubion, W.A.; Crotty, S.; McKean, D.J. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J. Immunol. 2006, 176, 291–300.
- Tembhare, P.R.; Ghogale, S.; Tauro, W.; Badrinath, Y.; Deshpande, N.; Kedia, S.; Cherian, K.; Patkar, N.V.; Chatterjee, G.; Gujral, S.; et al. Evaluation of CD229 as a new alternative plasma cell gating marker in the flow cytometric immunophenotyping of monoclonal gammopathies. Cytom. Part B Clin. Cytom. 2018, 94, 509–519.
- Yousef, S.; Kovacsovics-Bankowski, M.; Salama, M.E.; Bhardwaj, N.; Steinbach, M.; Langemo, A.; Kovacsovics, T.; Marvin, J.; Binder, M.; Panse, J.; et al. CD229 is expressed on the surface of plasma cells carrying an aberrant phenotype and chemotherapy-resistant precursor cells in multiple myeloma. Hum. Vaccines Immunother. 2015, 11, 1606–1611.
- Ishibashi, M.; Takahashi, R.; Tsubota, A.; Sasaki, M.; Handa, H.; Imai, Y.; Tanaka, N.; Tsukune, Y.; Tanosaki, S.; Ito, S.; et al. SLAMF3-Mediated Signaling via ERK Pathway Activation Promotes Aggressive Phenotypic Behaviors in Multiple Myeloma. Mol. Cancer Res. MCR 2020, 18, 632–643.
- Pojero, F.; Flores-Montero, J.; Sanoja, L.; Pérez, J.J.; Puig, N.; Paiva, B.; Bottcher, S.; van Dongen, J.J.; Orfao, A. Utility of CD54, CD229, and CD319 for the identification of plasma cells in patients with clonal plasma cell diseases. Cytom. Part B Clin. Cytom. 2016, 90, 91–100.
- Ijaz, M.; Wang, F.; Shahbaz, M.; Jiang, W.; Fathy, A.H.; Nesa, E.U. The Role of Grb2 in Cancer and Peptides as Grb2 Antagonists. Protein Pept. Lett. 2018, 24, 1084–1095.
- Lin, Q.; Zhao, J.; Song, Y.; Liu, D. Recent updates on CAR T clinical trials for multiple myeloma. Mol. Cancer 2019, 18, 154.
- Cunninghame-Graham, D.S.; Vyse, T.J.; Fortin, P.R.; Montpetit, A.; Cai, Y.C.; Lim, S.; McKenzie, T.; Farwell, L.; Rhodes, B.; Chad, L.; et al. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008, 9, 93–102.
- Margraf, S.; Garner, L.I.; Wilson, T.J.; Brown, M.H. A polymorphism in a phosphotyrosine signalling motif of CD229 (Ly9, SLAMF3) alters SH2 domain binding and T-cell activation. Immunology 2015, 146, 392–400.
- Ishibashi, M.; Sunakawa-Kii, M.; Kaito, Y.; Kinoshita, R.; Asayama, T.; Kuribayashi, Y.; Inokuchi, K.; Morita, R.; Tamura, H. The SLAMF3 rs509749 polymorphism correlates with malignant potential in multiple myeloma. Exp. Hematol. 2020.
- Radhakrishnan, S.V.; Luetkens, T.; Scherer, S.D.; Davis, P.; Vander-Mause, E.R.; Olson, M.L.; Yousef, S.; Panse, J.; Abdiche, Y.; Li, K.D.; et al. CD229 CAR T cells eliminate multiple myeloma and tumor propagating cells without fratricide. Nat. Commun. 2020, 11, 798.





