
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ani-Simona Sevastre | + 5034 word(s) | 5034 | 2021-01-15 08:44:12 | | | |
| 2 | Dean Liu | -2218 word(s) | 2816 | 2021-01-27 10:24:03 | | |
Video Upload Options
Epidermal growth factor receptor (EGFR) amplification is a characteristic of the classical subtype of glioma.
1. Introduction
With an overall survival of less than 35% in five years[1], malignant primary brain tumors are the most difficult to treat cancers. Of those, the most common type is represented by gliomas. Based on the expression patterns’ differences, glioblastomas are divided into three subtypes as follows: classical, proneural, and mesenchymal[2]. Because glioblastoma multiforme (GBM), a grade IV glioma[3], is one of the most aggressive primary brain tumors, recent studies and reviews have focused on deepening our understanding of the disease[4][5][6][7][8][9].
At present, GBM’s pathogenesis is better understood due to recent advances in molecular biology. For newly diagnosed glioblastoma, the current standard of care is represented by resection, followed by radiotherapy and temozolomide (TMZ) administration[10], but the median overall survival (OS) is not fully improved; therefore, new diagnosis and treatment strategies are needed[11][12].
Glioblastoma is the most common and the most deleterious glioma[13]. The 2011–2015 Statistical Report of the Central Brain Tumor Registry of the United States (CBTRUS) showed that glioblastoma represents 48% of the malignant brain and central nervous system tumors, with an incidence rate in the United States 1.58 times higher in males compared to females[14]. Due to the quick progression, even with aggressive multimodal treatment, glioblastoma remains almost incurable[15][16].
Nowadays, chemotherapy has a significant role in glioblastoma’s treatment strategies, with numerous research studies aimed to develop more efficient chemotherapeutic drugs[17]. Understanding the disease’s pathogenesis has a key role in identifying disease biomarkers and developing new potential chemotherapeutic drugs. We present some of the most promising signaling pathways involved in pathogenesis, with their specific targeting components.
GBM is characterized by nuclear atypia, cellular pleomorphism, mitotic activity, anaplasia, and rapid proliferation alternated with an aggressive invasion of the surrounding brain tissue. In its microenvironment, glioma cells are faced with many challenges such as acidity, hypoxia, and low nutrient availability. To maintain rapid growth, they need to modulate metabolic activity[18][19].
In multicellular organisms, tyrosine phosphorylation is involved in signal transduction, leading to differentiation, proliferation, migration, and survival[20][21].
Receptor tyrosine kinases (RTKs) are activated by binding their extracellular domain to corresponding ligands determining their oligomerization. This process activates the intracellular domain, facilitating the recruitment of proteins that start a signaling cascade, integrating numerous signaling pathways that lead to specific cellular responses[22]. Among all RTKs, epidermal growth factor receptor (EGFR) is the most amplified in GBM [23]. EGFR amplification is observed in the classical subtype of glioma[2]. EGFR gene amplification is detected in 57.4% of primary GBM patients, leading to high levels of EGFR protein, contributing to tumorigenesis and progression[24].
However, targeted therapies against this type of receptor have not yet shown a clear clinical benefit. Many factors contribute to resistance, such as ineffective blood–brain barrier penetration, heterogeneity, mutations, and compensatory signaling pathways.
2. Understanding EGFR Features
The transmembrane receptor of tyrosine kinase epidermal growth factor (EGFR), also known as HER (human EGFR related) 1 or ErbB1, along with HER2/neu (ErbB-2), HER3 (ErbB-3), and HER4 (ErbB-4), is a member of the ErbB family and it is located on chromosome band 7p12 [25].
Like all RTKs, EGFR has an extracellular region, a single transmembrane domain, an intracellular juxtamembrane domain, a tyrosine kinase, and a C-terminal region. The ligands of ErbB receptors are divided into two main groups: EGFR activators called EGF agonists, and neuregulins that bind to ErbB3 and ErbB4[26][27].
The extracellular region of EGFR has two homologous domains (I and III) that bind ligands and two cysteine rich domains (II and IV)[28].
The juxtamembrane region tethers inactive EGFRs to the plasma membrane cytosolic surface, which contributes to EGFR activation[29]. Structural studies highlight the functional importance for certain regions, such as the structure of the first 30 amino acids from the intracellular juxtamembrane region of EGFR and the C-terminal 190 amino acids [27].
There are more than 40 EGFR ligands that control its signaling. They can be divided into high-affinity ligands, such as epithelial growth factor (EGF), heparin-binding EGF-like growth factor (HB-EGF), Transforming growth factor alpha (TGF-α), betacellulin (BTC), and low-affinity ligands, such as epiregulin (EREG), amphiregulin (AREG), and epigen (EPGN).
Expression of EGF-family proteins and activation of EGFR are features of cardiac disease[30][31]. Moreover, molecular alterations of EGFR include overexpression, deletion, or amplification, in different types of cancer. In GBM, EGFR amplification promotes invasion, proliferation, and drug resistance to radio- and chemotherapy[32].
Several trials on EGFR targeted therapy have failed to produce conclusive evidence, maybe because of the EGFR molecular heterogeneity in GBM, of the low specificity of the designed drugs, as well as because of low brain penetration[33]. Despite all this, the detection of EGFR alterations is still used as a prognostic marker for GBM because 24–67% of GBMs are characterized by a mutated gene, 40% by amplification, and 60% by EGFR overexpression[34].
In recent years, studies have proved that EGFR has pro-survival kinase-independent functions in malignant cells. This fact has offered a different perspective of understanding EGFR implications in cancer, with new ideas of EGFR targeted cancer therapy[35][36][37].
3. Mechanisms of EGFR Pathway Activation
There are several different mechanisms of EGFR pathway activation, such as increased ligand production or overexpression/defective inactivation/mutation of the receptor. Many studies focused on the EGFR signaling mechanism in recent years, trying to conclude how the extracellular EGFR-ligand binding propagates through the single transmembrane helix (TM) to trigger intracellular kinase activation[38][39][40].
3.1. EGFR Activation Mechanisms in Normal Physiologic Status
The expression of EGFR in normal cells is about 4 × 104–10 × 104 receptors/cell[41] whereas, in cancer cells, more than 106 receptors/cell are observed[42]
The EGFR RNA expression is increased by stimulating the EGFR-specific transcription factor (ETF). The receptor expression is regulated by epidermal growth factor (EGF) itself and other proteins such as E1A, Sp1, and AP2[36].
Like all RTKs, EGFR is activated by ligands featuring receptor-specificity. Briefly, ligand binding leads to a dimeric active conformation of EGFR by homodimerization (complexed with another EGFR) or heterodimerization (complex with another ErbB member). The tyrosine residues from other RTKs are autophosphorylated after ligand stimulation, and phenylalanine substitutions significantly impair the kinase signaling and the downstream signaling. Differently, EGFR Tyr-845 phosphorylation is not a required mechanism for ligand-induced EGFR activation, but it may represent the main mechanism for EGFR transactivation[43][44].
Proteins that express a proto-oncogene tyrosine-protein kinase (Src) homology domain 2 (SH2) region bind to the activated receptor, areactivated, and forward the signal to the downstream effectors, propagating critical cellular signaling pathways[45]. EGFR can simultaneously activate several signal transduction pathways such as phosphatidylinositide 3 kinase (PI3K) and serine–threonine kinase (AKT) and RAS/MAPK pathways[46].
3.1.1. Extracellular Domain Activation
For EGFR, the dimerization is completely receptor-mediated, with no physical interaction between two activating ligands. In normal physiologic status, the receptors are in a dynamic monomer–dimer equilibrium. In the absence of ligands, the extracellular domain presents a tethered configuration (intra-molecular links entirely block the dimerization arm), and the intracellular tyrosine kinase domain (TKD) is inactive. Ligand binding leads to a conformational change that exposes the buried dimerization arm, and the extracellular domain dimerizes, inducing conformational changes of the intracellular domain and enabling kinase activation[45].
A recent study by Chung et al. described physiological EGFR activation as being due to a ligand-mediated extracellular domain dimerization that stabilizes the N-terminal transmembrane dimer and disrupts autoinhibition, allowing the C-terminal juxtamembrane (JM-B) segment to stabilize the asymmetric kinase domain (KD) dimer, resulting in activation of EGFR signaling. They also concluded that the stimulus stabilizes the active KD conformation in pathological states and further the asymmetric KD dimerization. The inside–out coupling is weaker than the physiological outside–in coupling, suggesting that the extracellular (EC) dimer is linked through the N-terminal TM dimer with the asymmetric oncogenic KD dimer [38].
3.1.2. Intracellular Domains Activation
By ligand-induced dimerization, the cis-autoinhibition is released, and through a unique allosteric mechanism, the kinase activity of EGFR isactivated. It is well known that this mechanism consists of physical interaction between the C-terminal tail of the activator kinase and the other kinase N-terminal tail (receiver kinase) of the dimer pair, inducing conformational changes of the N-lobe of receiver kinase and trans-phosphorylation C-terminal tail of the activator [47].
3.1.3. Downstream Signaling of EGFR
EGFR activation and autophosphorylation result in the recruitment of downstream signaling proteins. Almost all autophosphorylation sites are binding sites for Src Homology 2-(SH2) or Phosphotyrosine binding-(PTB) signaling proteins. The SH2- proteins may be bound directly to the receptor, or indirectly through docking proteins using PTB domains[48]. EGFR can recruit and regulate many signaling pathways such as PI-3 K/AKT, RAS/MAPK, and JAK2/STAT. Therefore, EGFR functions as a hub involved in regulating various cellular processes[21][23], as shown in Figure 1.
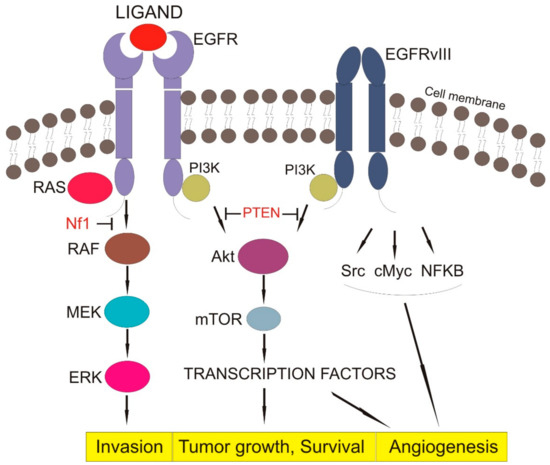
Figure 1. EGFR signaling pathway (EGFR—epithelial growth factor receptor, EGFRvIII—Epidermal growth factor receptor variant III, Pi3K—Phosphoinositide 3-kinase, RAS—family of genes involving cellular signal transduction, PTEN—Phosphatase and tensin homolog, NF1—Neurofibromatosis type 1, RAF—serine/threonine-specific protein kinases, MEK—Mitogen-activated protein kinase, ERK—extracellular signal-regulated kinase, AkT—Protein kinase B, mTOR—mammalian target of rapamycin, Src—Proto-oncogene tyrosine-protein kinase, cMyc—c proto-oncogene, NFKB—nuclear factor kappa-light-chain-enhancer of activated B cells, Block arrow—inhibition activity, Point arrow—pathway flow).
The PI-3K/AKT signaling pathway involves PI3K, an enzyme with SH2-signal transducer and its downstream effector AKT, regulating apoptosis and cell survival. Once EGFR is activated and phosphorylated, PI3K is brought to the cell membrane, and it phosphorylates phosphatidylinositol 4,5-bisphosphate (PIP2), forming phosphatidylinositol (3,4,5)-trisphosphate (PIP3). AKT reacts with PIP3, and it is phosphorylated at Threonin308 by phosphoinosite-dependent protein kinase-1 (PDK1) and at Serine 473 by the mammalian target of rapamycin complex 2 (mTORC2), reaching full activity. The phosphatase and tensin homolog (PTEN) negatively regulate the PI3K/AKT pathway by dephosphorylating and delocalizing PIP3 from the cellular membrane, resulting in the relocalization of AKT in the cytoplasm, where it is unable to be reactivated[49][50].
Class IA is one of the three different classes of PI3Ks featuring subunits with regulatory activity such as p85. Active EGFR achieves association with regulatory p85 through dimerization with human HER3, or via the docking protein GRB2-associated binder 1 (GAB1), relieving the inhibitory effect of p85[51]. GAB1 is a scaffolding protein involved in recruiting additional signaling proteins such as PI3K, SHP2, and p120RasGap. It is involved in many EGFR signaling outputs, and is the predominant mechanism linking EGFR to PI3K/Akt signaling[52][53].
Due to its increasing importance in different human cancers, GAB1 may represent an emerging potential therapeutic target.
The RAS/MAPK signaling pathway involves the growth-factor-receptor bound-2 (GRB2), which forms a complex with Son of Sevenless (SOS), a guanine-nucleotide exchange factor (GEF) and activates the RAS G-protein by exchanging guanosine diphosphate (GDP) with guanosine triphosphate (GTP)[54]. Consequently, RAS and mitogen-activated protein kinases (MAPKs) initiate a downstream signaling cascade to phosphorylate the nuclear protein Jun. Jun creates complexes with different nuclear proteins leading to the key transcription factor activator protein 1 (AP-1), responsible for translation and transcription of proteins involved in the growth and division of cells. Activated RAS is negatively regulated by GTPase activating proteins (GAPs), such as the tumor suppressor neurofibromin 1 (NF1) [55].
Signal transduction and activator of transcription 3 (STAT3) is tyrosine-phosphorylated or activated as pSTAT3 due to EGFR-regulation of interleukin-6 (IL-6) expression. This mechanism leads to a feed-forward in the IL-6/Janus kinase (JAK)/STAT3 loop [21][56][57][58].
3.2. Oncogenic Status and EGFR Activation
The EGFR is one of the most frequently altered oncogenes in brain cancers. Except for hematopoietic cells, the majority of cell types express ErbB family members[35].
In glioblastoma cells, the EGFR tyrosine kinase activity may be dysregulated by multiple oncogenic mechanisms, such as gene mutation, overexpression of EGFR protein, increased gene copy number, rearrangements of chromosomes, and activation by autocrine function[59].
3.2.1. Mutations of Cell Signaling Regulators
The EGFR gene is located on chromosome 7p11.2 and consists of 28 exons encoding a transmembrane protein receptor composed of 464 amino acids. Exons 5–7 and 13–16 encode the ligand binding domain, and exons 18–24 encode the tyrosine kinase domain. The region encoded by exons 25–28 is the site of autophosphorylation.
Although EGFR is one of the most important drug targets in cancer therapies, its mutations present an organ–site asymmetry, depending on the cancer’s organ of origin[60]. Although mutations occur in the kinase domain (KD) in other tumors, in gliomas, heterogeneous mutations and deletions are focused on the ligand-binding ectodomain (ECD). This tissue-specific feature leads to type-II tyrosine kinase inhibitors (TKIs) with high sensitivity for the inactive symmetric KD dimer (sKD), when administered in GBM mutations[61]. However, both intra- and extracellular GBM mutations result in ligand-independent oncogenic activation.
Almost 50% of the tumors characterized by EGFR amplification are positive for the mutant EGFRvIII and EGFR single nucleotide variants (SNVs). Due to this tumor-specific feature, novel therapeutic agents are currently under development to target the overexpressed EGFR or EGFRvIII proteins. An in-frame deletion of exons 2–7 characterizes the EGFRvIII, which results in overexpression of a truncated receptor that lacks some significant parts of the ECD. This prototypic oncoprotein is unable to bind ligands, and it is constitutively active. Several studies examined the effect of the EGFRvIII constitutive activity on the wtEGFR and ErbB2 protein levels. For example, one study evaluated the effect of Tyrphostin AG1478 on the protein levels and demonstrated that its administration increased protein levels of wtEGFR and erbB2 in vIIIA1 cells, due to the catalytic activity of EGFRvIII, while in its absence, the levels were reduced[62]. Furthermore, the unique peptide sequence of EGFRvIII generated by the fusion of exons 1 and 8 may serve as a tumor-specific target in immunotherapy[63], although subsequent phase III trial results are not as promising as initially anticipated[64].
A meta-analysis performed in 2017 by Felsberg et al. proved that EGFRvIII and EGFR SNVs do not represent prognostic keys in EGFR-amplified glioma patients. However, the amplification of EGFR is retained in recurrent glioma[63], although improved long-term survival by EGFRvIII therapy has been reported in glioblastoma patients [65].
Nevertheless, the research on EGFRvIII continues, producing inconclusive results. For example, Struve et al. just published in early 2020 the results of a study focused on the effect of EGFRvIII in regulating DNA mismatch repair. They tested if EGFRvIII influences temozolomide’s sensitivity and demonstrated that, under standard treatment with temozolomide, EGFRvIII expression leads to prolonged survival only in patients with tumors with O6-methylguanine-DNA methyltransferase (MGMT) methylated promoter. Their results showed that EGFRvIII sensitizes a type of GBM to the current standard of care treatment with temozolomide through the upregulation of DNA mismatch repair (MMR) [65]. However, patients with tumors that have both EGFRvIII and MGMT methylation are very uncommon, and the conclusion that EGFRvIII status was associated with increased survival had a p = 0.06. This level would not normally be considered significant, especially not in this sort of multivariate analysis[66].
3.2.2. Overexpression and Gene Amplification
The EGFR gene is amplified in approximately 40% of glioblastomas. The primary and secondary GBM differ in genetic profiles and primary GBMs have a higher prevalence of EGFR gene amplification and overexpression than secondary GBMs[67]. In a study performed by Watanabe et al., EGFR gene amplification was associated with protein overexpression in most tumor cells, but 10% of GBM with overexpression of EGFR protein lacked EGFR gene amplification[68]. However, previous studies have stated that EGFR overexpression or activation does not necessarily cause a simple amplification of its downstream signals, but dose-dependent changes in oncogene-induced downstream signaling and biological responses have been reported [69].
3.2.3. Rearrangements of Chromosomes
Breakpoint sequence analyses proved different types of chromosomal rearrangements and mechanisms of DNA repair. Analyses of single nucleotide polymorphisms suggested that different deletions may appear from amplified non-vIII EGFR precursor[70].
In a study performed in 2018 on glioma tumor samples by Tomoyuki et al., complex chromosomal rearrangements involving chromosome 7 were observed[70].
A study performed by Lopez-Gines et al. showed that trisomy/polysomy 7 and monosomy 10 were frequently associated with glioma. The combination of these anomalies is important in glioblastoma’s tumorigenesis. Moreover, the association seems to be independent of EGFR gene amplification[71].
3.2.4. Activation by Autocrine Function
It is well known that wild-type EGFR ligands such as transforming growth factor-alpha (TGF-alpha) and heparin-binding EGF (HB-EGF) are often increased in glioblastoma leading to an autocrine loop resulting in the autonomy growth of glioma cells[72]. GBM expresses an EGFR mutant (EGFRvIII) that signals constitutively, does not bind ligand, and is considered to have more tumorigenicity than wild-type EGFR. In a U251-MG glioma cell line, the expression of EGFRvIII may result in specific up-regulation of some genes (TGF-α, EPHA2, HB-EGF, IL8, FOSL1, MAP4K4, DUSP6, and EMP1) influencing signaling pathways involved in oncogenesis. TGF-α and HB-EGF (EGFR ligands) induce the expression of EGFRvIII, suggesting that EGFRvIII has a role in creating an autocrine loop with wild-type EGFR. By inhibiting HB-EGF activity with neutralizing antibodies, EGFRvIII-induced cell proliferation may be reduced, suggesting that EGFRvIII-HB-EGF-wild-type EGFR autocrine loop has a major role in signal transduction in glioblastoma cells[73]. Furthermore, studies have demonstrated that the expression of the EGFR alone has a poor transformation effect on cells. Though, coexpression of TGF-α ligand leads to a significant increase in transformation and therapies based on neutralizing the ligands have demonstrated the decreased growth of cells that harbor such loops[74][75].
References
- Zhang, A.S.; Ostrom, Q.T.; Kruchko, C.; Rogers, L.; Peereboom, D.M.; Barnholtz-Sloan, J.S. Complete prevalence of malignant primary brain tumors registry data in the United States compared with other common cancers 2010. Neuro Oncol. 2017, 19, 726–735.
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010, 17, 98–110.
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820.
- Armstrong, T.S.; Dirven, L.; Arons, D.; Bates, A.; Chang, S.M.; Coens, C.; Espinasse, C.; Gilbert, M.R.; Jenkinson, D.; Kluetz, P.; et al. Glioma patient-reported outcome assessment in clinical care and research: A Response Assessment in Neuro-Oncology collaborative report. Lancet Oncol. 2020, 21, e97–e103.
- Alexandru, O.; Sevastre, A.S.; Castro, J.; Artene, S.A.; Tache, D.E.; Purcaru, O.S.; Sfredel, V.; Tataranu, L.G.; Dricu, A. Platelet-Derived Growth Factor Receptor and Ionizing Radiation in High Grade Glioma Cell Lines. Int. J. Mol. Sci. 2019, 20, 4663.
- Alexandru, O.; Horescu, C.; Sevastre, A.S.; Cioc, C.E.; Baloi, C.; Oprita, A.; Dricu, A. Receptor tyrosine kinase targeting in glioblastoma: Performance, limitations and future approaches. Współczesna Onkol. 2020, 24, 55–66.
- Sevastre, A.-S.; Horescu, C.; Carina Baloi, S.; Cioc, C.E.; Vatu, B.I.; Tuta, C.; Artene, S.A.; Danciulescu, M.M.; Tudorache, S.; Dricu, A. Benefits of Nanomedicine for Therapeutic Intervention in Malignant Diseases. Coatings 2019, 9, 628.
- Novak, M.; KoprivnikarKrajnc, M.; Hrastar, B.; Breznik, B.; Majc, B.; Mlinar, M.; Rotter, A.; Porčnik, A.; Mlakar, J.; Stare, K.; et al. CCR5-Mediated Signaling Is Involved in Invasion of Glioblastoma Cells in Its Microenvironment. Int. J. Mol. Sci. 2020, 21, 4199.
- Alexandru, O.; Dragutescu, L.; Tătăranu, L.; Ciubotaru, V.; Sevastre, A.; Georgescu, A.M.; Purcaru, O.; Dănoiu, S.; Bäcklund, L.M.; Dricu, A. Helianthin induces antiproliferative effect on human glioblastoma cells in vitro. J. Neuro Oncol. 2011, 102, 9–18.
- Fernandes, C.; Costa, A.; Osório, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Glioblastoma [Internet]. Current Standards of Care in Glioblastoma Therapy; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017; Chapter 11. Available online: https://www.ncbi.nlm.nih.gov/books/NBK469987/ (accessed on 22 June 2020).
- Rajaratnam, V.; Islam, M.M.; Yang, M.; Slaby, R.; Ramirez, H.M.; Mirza, S.P. Glioblastoma: Pathogenesis and Current Status of Chemotherapy and Other Novel Treatments. Cancers 2020, 12, 937.
- Horescu, C.; Cioc, C.E.; Tuta, C.; Sevastre, A.S.; Tache, D.E.; Alexandru, O.; Artene, S.A.; Danoiu, S.; Dricu, A.; Purcaru, S.O. The effect of temozolomide in combination with doxorubicin in glioblastoma cells in vitro. J. Immunoass. Immunochem. 2020, 6, 1–11.
- Stöppler, M.C.; Shiel, W.C.; Credo Reference (Firm); WebMD (Firm). Webster’s New World Medical Dictionary, 3rd ed.; Credo Reference: Boston, MA, USA; Wiley: Hoboken, NJ, USA, 2014; p. 1.
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol. 2018, 20, iv1–iv86.
- Tirosh, I.; Suvà, M.L. Tackling the Many Facets of Glioblastoma Heterogeneity. Cell Stem Cell. 2020, 26, 303–304.
- Carapancea, M.; Alexandru, O.; Fetea, A.S.; Dragutescu, L.; Castro, J.; Georgescu, A.; Popa-Wagner, A.; Bäcklund, M.L.; Lewensohn, R.; Dricu, A. Growth factor receptors signaling in glioblastoma cells: Therapeutic implications. J. Neurooncol. 2009, 92, 137–147.
- Oprita, A.; Sevastre, A.S. New pharmaceutical dosage forms used in the treatment of breast cancer. Polymeric micelles. Med. Oncol. 2020, 1, 38–52.
- Mao, H.; Lebrun, D.G.; Yang, J.; Zhu, V.F.; Li, M. Deregulated signaling pathways in glioblastoma multiforme: Molecular mechanisms and therapeutic targets. Cancer Investig. 2012, 30, 48–56.
- Alexandru, O.; Ciubotaru, V.; Tataranu, L.; Fetea, S.; Badea, P.; Dricu, A. The relationship between cognitive function, tumour histology and surgical treatment in patients with primary brain tumours. Communications of the European Neurological Society. J. Neurol. 2008, 255, 151–156.
- Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225.
- Eskilsson, E.; Røsland, G.V.; Solecki, G.; Wang, Q.; Harter, P.N.; Graziani, G.; Verhaak, R.G.W.; Winkler, F.; Bjerkvig, R.; Miletic, H. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol. 2018, 20, 743–752.
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146.
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575.
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477.
- Zhang, H.; Berezov, A.; Wang, Q.; Zhang, G.; Drebin, J.; Murali, R.; Greene, M.I. ErbB receptors: From oncogenes to targeted cancer therapies. J. Clin. Investig. 2007, 117, 2051–2058.
- Wood, E.R.; Truesdale, A.T.; McDonald, O.B.; Yuan, D.; Hassell, A.; Dickerson, S.H.; Ellis, B.; Pennisi, C.; Horne, E.; Lackey, K.; et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004, 64, 6652–6659.
- Ferguson, K.M. Structure-based view of epidermal growth factor receptor regulation. Annu. Rev. Biophys. 2008, 37, 353–373.
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398.
- Arkhipov, A.; Shan, Y.; Das, R.; Endres, N.; Eastwood, M.; Wemmer, D.; Kuriyan, J.; Shaw, D. Architecture and Membrane Interactions of the EGF Receptor. Cell 2013, 152, 557–569.
- Forrester, S.J.; Kawai, T.; O’Brien, S.; Thomas, W.; Harris, R.C.; Eguchi, S. Epidermal Growth Factor Receptor Transactivation: Mechanisms, Pathophysiology, and Potential Therapies in the Cardiovascular System. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 627–653.
- Makki, N.; Thiel, K.W.; Miller, F.J., Jr. The epidermal growth factor receptor and its ligands in cardiovascular disease. Int. J. Mol. Sci. 2013, 14, 20597–20613.
- Alexandru, O.; Purcaru, S.O.; Tataranu, L.G.; Lucan, L.; Castro, J.; Folcuţi, C.; Artene, S.A.; Tuţă, C.; Dricu, A. The Influence of EGFR Inactivation on the Radiation Response in High Grade Glioma. Int. J. Mol. Sci. 2018, 19, 229.
- Kwatra, M.M. A Rational Approach to Target the Epidermal Growth Factor Receptor in Glioblastoma. Curr. Cancer Drug Targets 2017, 17, 290–296.
- Muñoz-Hidalgo, L.; San-Miguel, T.; Megías, J.; Monleón, D.; Navarro, L.; Roldán, P.; Cerdá-Nicolás, M.; López-Ginés, C. Somatic copy number alterations are associated with EGFR amplification and shortened survival in patients with primary glioblastoma. Neoplasia 2020, 22, 10–21.
- Thomas, R.; Weihua, Z. Rethink of EGFR in Cancer with Its Kinase Independent Function on Board. Front. Oncol. 2019, 9, 800.
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20.
- Jureczek, J.; Feldmann, A.; Bergmann, R.; Arndt, C.; Berndt, N.; Koristka, S.; Loureiro, L.R.; Mitwasi, N.; Hoffmann, A.; Kegler, A.; et al. Highly Efficient Targeting of EGFR-Expressing Tumor Cells with UniCAR T Cells via Target Modules Based on Cetuximab®. Onco Targets Ther. 2020, 13, 5515–5527.
- Tsai, C.-J.; Nussinov, R. Emerging Allosteric Mechanism of EGFR Activation in Physiological and Pathological Contexts. Biophys. J. 2019, 117, 5–13.
- Hsu, P.C.; Jablons, D.M.; Yang, C.T.; You, L. Epidermal Growth Factor Receptor (EGFR) Pathway, Yes-Associated Protein (YAP) and the Regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC). Int. J. Mol. Sci. 2019, 20, 3821.
- Vitiello, P.P.; Cardone, C.; Martini, G.; Ciardiello, D.; Belli, V.; Matrone, N.; Barra, G.; Napolitano, S.; Della Corte, C.; Turano, M.; et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J. Exp. Clin. Cancer Res. 2019, 38, 41.
- Carpenter, G.; Cohen, S. Epidermal growth factor. Annu. Rev. Biochem. 1979, 48, 193–216.
- Gullick, W.J.; Marsden, J.J.; Whittle, N.; Ward, B.; Bobrow, L.; Waterfield, M.D. Expression of Epidermal Growth Factor Receptors on Human Cervical, Ovarian, and Vulval Carcinomas. Cancer Res. 1986, 46, 285–292.
- Parsons, J.T.; Parsons, S.J. Src family protein tyrosine kinases: Cooperating with growth factor and adhesion signaling pathways. Curr. Opin. Cell Biol. 1997, 9, 187–192.
- Biscardi, J.S.; Maa, M.C.; Tice, D.A.; Cox, M.E.; Leu, T.H.; Parsons, S.J. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 1999, 274, 8335–8343.
- Dawson, J.P.; Berger, M.B.; Lin, C.C.; Schlessinger, J.; Lemmon, M.A.; Ferguson, K.M. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell. Biol. 2005, 25, 7734–7742.
- Hervieu, A.; Kermorgant, S. The Role of PI3K in Met Driven Cancer: A Recap. Front. Mol. Biosci. 2018, 5, 86.
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134.
- Brummer, T.; Schmitz-Peiffer, C.; Daly, R.J. Docking proteins. FEBS J. 2010, 277, 4356–4369.
- Kharbanda, A.; Walter, D.; Gudiel, A.; Schek, N.; Feldser, D.; Witze, E. Blocking EGFR palmitoylation suppresses PI3K signaling and mutant KRAS lung tumorigenesis. Sci. Signal. 2020, 13, eaax2364.
- Mayer, I.A.; Arteaga, C.L. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu. Rev. Med. 2016, 67, 11–28.
- Mattoon, D.R.; Lamothe, B.; Lax, I.; Schlessinger, J. The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol. 2004, 2, 24.
- Mulcahy, E.Q.X.; Colόn, R.R.; Abounader, R. HGF/MET Signaling in Malignant Brain Tumors. Int. J. Mol. Sci. 2020, 21, 7546.
- Kiyatkin, A.; Aksamitiene, E.; Markevich, N.I.; Borisov, N.M.; Hoek, J.B.; Kholodenko, B.N. Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J. Biol. Chem. 2006, 281, 19925–19938.
- Pawson, T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 2004, 116, 191–203.
- Ward, A.F.; Braun, B.S.; Shannon, K.M. Targeting oncogenic Ras signaling in hematologic malignancies. Blood 2012, 120, 3397–3406.
- Gao, S.P.; Mark, K.G.; Leslie, K.; Pao, W.; Motoi, N.; Gerald, W.L.; Travis, W.D.; Bornmann, W.; Veach, D.; Clarkson, B.; et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Investig. 2007, 117, 3846–3856.
- Gao, S.P.; Chang, Q.; Mao, N.; Daly, L.A.; Vogel, R.; Chan, T.; Liu, S.H.; Bournazou, E.; Schori, E.; Zhang, H.; et al. JAK2 inhibition sensitizes resistant EGFR-mutant lung adenocarcinoma to tyrosine kinase inhibitors. Sci. Signal. 2016, 9, ra33.
- Padfield, E.; Ellis, H.P.; Kurian, K.M. Current Therapeutic Advances Targeting EGFR and EGFRvIII in Glioblastoma. Front. Oncol. 2015, 5, 5.
- Jorissen, R.N.; Walker, F.; Pouliot, N.; Garrett, T.P.; Ward, C.W.; Burgess, A.W. Epidermal growth factor receptor: Mechanisms of activation and signalling. Exp. Cell Res. 2003, 284, 31–53.
- Huang, P.H.; Xu, A.M.; White, F.M. Oncogenic EGFR signaling networks in Glioma. Sci. Signal. 2009, 2, 1–13.
- Furnari, F.B.; Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat. Rev. Cancer 2015, 15, 302–310.
- Zeineldin, R.; Ning, Y.; Hudson, L.G. The constitutive activity of epidermal growth factor receptor vIII leads to activation and differential trafficking of wild-type epidermal growth factor receptor and erbB2. J. Histochem. Cytochem. 2010, 58, 529–541.
- Felsberg, J.; Hentschel, B.; Kaulich, K.; Gramatzki, D.; Zacher, A.; Malzkorn, B.; Kamp, M.; Sabel, M.; Simon, M.; Westphal, M.; et al. Epidermal Growth Factor Receptor Variant III (EGFRvIII) Positivity in EGFR-Amplified Glioblastomas: Prognostic Role and Comparison between Primary and Recurrent Tumors. Clin. Cancer Res. 2017, 23, 6846–6855.
- Platten, M. EGFRvIII vaccine in glioblastoma-InACT-IVe or not ReACTive enough? Neuro Oncol. 2017, 19, 1425–1426.
- Struve, N.; Binder, Z.A.; Stead, L.F.; Brend, T.; Bagley, S.J.; Faulkner, C.; Ott, L.; Müller-Goebel, J.; Weik, A.-S.; Hoffer, K.; et al. EGFRvIII upregulates DNA mismatch repair resulting in increased temozolomide sensitivity of MGMT promoter methylated glioblastoma. Oncogene 2020, 39, 3041–3055.
- Brito, C.; Azevedo, A.; Esteves, S.; Marques, A.R.; Martins, C.; Costa, I.; Mafra, M.; Bravo Marques, J.M.; Roque, L.; Pojo, M. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer 2019, 19, 968.
- Ohgaki, H.; Kleihues, P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453.
- Watanabe, K.; Tachibana, O.; Sata, K.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996, 6, 217–223.
- Sarkisian, C.J.; Keister, B.A.; Stairs, D.B.; Boxer, R.B.; Moody, S.E.; Chodosh, L.A. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat. Cell Biol. 2007, 9, 493–505.
- Koga, T.; Li, B.; Figueroa, J.M.; Ren, B.; Chen, C.C.; Carter, B.S.; Furnari, F.B. Mapping of genomic EGFRvIII deletions in glioblastoma: Insight into rearrangement mechanisms and biomarker development. Neuro Oncol. 2018, 20, 1310–1320.
- Lopez-Gines, C.; Cerda-Nicolas, M.; Gil-Benso, R.; Pellin, A.; Lopez-Guerrero, J.A.; Callaghan, R.; Benito, R.; Roldan, P.; Piquer, J.; Llacer, J.; et al. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin. Neuropathol. 2005, 24, 209–218.
- Singh, B.; Carpenter, G.; Coffey, R.J. EGF receptor ligands: Recent advances. F1000Research 2016, 5, 2270.
- Ramnarain, D.B.; Park, S.; Lee, D.Y.; Hatanpaa, K.J.; Scoggin, S.O.; Out, H.; Libermann, T.A.; Raisanen, J.M.; Ashfaq, R.; Wong, E.T.; et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006, 66, 867–874.
- Tang, P.; Steck, P.A.; Yung, W.K. The autocrine loop of TGF-α/EGFR and brain tumors. J. Neurooncol. 1997, 35, 303–314.
- Filmus, J.; Shi, W.; Spencer, T. Role of transforming growth factor α (TGF-α) in the transformation of ras-transfected rat intestinal epithelial cells. Oncogene 1993, 8, 1017–1022.




