
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Abd-Elgawad Radi | + 3339 word(s) | 3339 | 2021-01-20 04:56:26 | | | |
| 2 | Dean Liu | -1625 word(s) | 1714 | 2021-01-22 03:34:06 | | | | |
| 3 | Dean Liu | -1625 word(s) | 1714 | 2021-01-22 03:38:24 | | | | |
| 4 | Catherine Yang | Meta information modification | 1714 | 2021-09-28 07:56:45 | | |
Video Upload Options
Over the last few decades, aptamers have attracted a lot of interest in the biosensor industry, because they are the next generation of target receptors that can replace antibody functions. SELEX is an automated procedure and needs only a few days to evolve some binders. This is much shorter compared to antibody selection, which often requires several months. Aptamers can even differentiate the chirality of a molecule and its secondary structure. Aptamers can choose any types of targets with no restrictions. The antibodies undergo permanent degradation, while aptamers can undergo several cycles of denaturation/regeneration. DNA aptamers are acceptable for the design of reusable aptamer detectors, while RNA aptamers can be single-dimensional. The use of aptamers is not limited to specific areas and can be used as recognition molecules in almost any domain. The main limitation is the degradation of RNAs aptamer by ribonuclease. These problems can be solved by modifying RNA aptamers. Another limitation is that the microenvironment will affect the structure of the aptamer and the interactions with the ligand-target. Moreover, the composition of salts has a significant effect on aptamer configuration. The integration of aptamers into detection platforms such as microfluidics and paper-based analytical devices and lab-on-a-chip (LOC) areas for point-of-care (POC) diagnosis is becoming increasingly popular. Aptamer-based detection systems meet most POC diagnostic requirements.
1. Introduction
The procedure for the in vitro selection of aptamers is called the systematic evolution of ligands by exponential enrichment (SELEX; Scheme 1)[1]. The first reports about SELEX were published by Gold’s and Szostak’s groups in 1990[2][3]. The SELEX launches with the creation of a multiple library of DNA or RNA molecules. The library is subsequently imported into a target ligand and the sequences, which reveal affinity towards the target molecule and are isolated from the unbound sequences. The bound sequences are then collected and subjected to PCR amplification for the following rounds of enrichment. Multiple rounds have been attained until the library reach to set of convergent sequences with an affinity for the target molecule. Since aptamers were found in the 1990s, they have attracted substantial attention and have been utilized in many fields[4]. The antibody-based platform can detect many targets, but it is always hampered by the antibody preparation time, antibody equilibrium, and the effect of the modification on the antibody. The monoclonal antibodies production is very expensive processes[5]. Moreover, immunoassays must be validated in every new batch. Seeking alternatives to antibodies is of enormous importance. Antibodies as proteins are easily denatured and shed their tertiary structure at high temperatures; however, oligonucleotides are more thermally stable and keep their structures over repeated cycles of denaturation/renaturation[6][7]. The aptamer modification by different chemical reactions improve their stability and nuclease resistance[8][9]. The aptamers may be directly chosen in vitro against different targets, from small molecules to proteins and even cells[10]. These advantages make it feasible for them to be used as a recognition probe to substitute antibodies in the diagnostic field. Great attention has been paid to these biosensors, due to their simple utility, accessibility, and real-time outputs. Therefore, sensors that use aptamers as their bio-recognition elements have found applications in diverse fields[10].
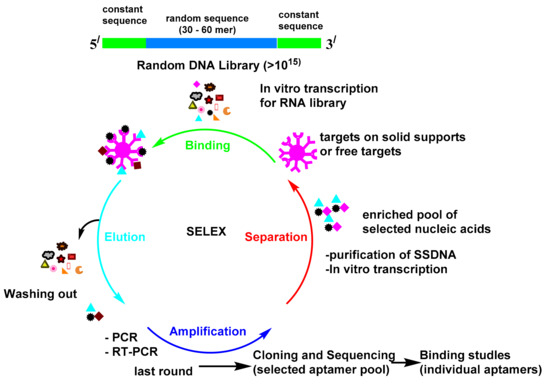
Scheme 1. Systematic evolution of ligands by exponential enrichment (SELEX).
A biosensor that is structured from aptamers as a recognition element is called an aptasensor. The electrochemical aptasensor was introduced for the first time in 2004[11]. Up to now, the best-investigated aptamers are those for thrombin[12][13][14][15][16]. A promising scope for the new era of diagnostics could be offered by the electrochemical biosensors. The principle of function depends on the change in the current, impedance, or potential, in response to the recognition event at the transducer of the sensor[17][18][19][20][21]. The electrochemical detection advantages are low-cost, fast, multi-analyte analyses, readily amenable for miniaturization, as well as being robust and compatible with novel microfabrication technologies and real time detection. These advantages offer the desired characteristics for point-of-care tests for monitoring of clinical analytes.
2. Electrochemical Aptasensors Configuration
The direct apta-assay formats are concerned with the molecular recognition of the target using the cognate aptamer immobilized onto the sensing interface. For example, a thiol terminated aptamer was immobilized on an Au electrode. After the thrombin (Tb) binding event, the Tb was detected by evaluation of the thrombin catalyzed reaction that produces p-nitroaniline[22]. The main disadvantage of such an approach is the high detection limits and the limited selectivity of the sensing layer. The biosensing assays could be improved using other different configurations; including sandwich-type, displacement-type and folding-based electrochemical aptasensors. This section discusses advances in the electrochemical aptasensors for target molecules monitoring, focusing on effective detection strategies for the formation of sandwich structures, and EIS measurement of the change of charge transfer resistance, which is related to the target molecule level, and the aptamer conformation change causing the electrochemical signal on/off of the electroactive markers.
2.1. Sandwich-Type Electrochemical Aptasensors
The sandwich-type construct involves antibody–aptamer sandwich sensing layers; the antibody is used as a capture element and aptamer as a reporter probe or vice versa. The signaling antibody is usually conjugated with a redox label, such as methylene blue (MB), ferrocene (Fc) or horseradish peroxidase (HRP) which can be detected using electrochemical techniques. [22][24].
The sandwich format generally offers higher selectivity as compared to single-receptor approaches, due to the involvement of a pair of receptors; the capture and reporter. In addition, the sandwich format promotes a wide variety of amplification strategies, which can be divided into two broad categories: depending on the nature of synthetic receptors or on the unique advantages of nanoparticles. In addition to DNA amplification, strategies based on the extension of the reporter aptamer with a proper primer thus provoke isothermal and linear RCA[25][26] nucleic acid nanomachines such as DNA walkers[27]. Scheme 2 shows the formation of sandwich-types structures for aptasensing assay.
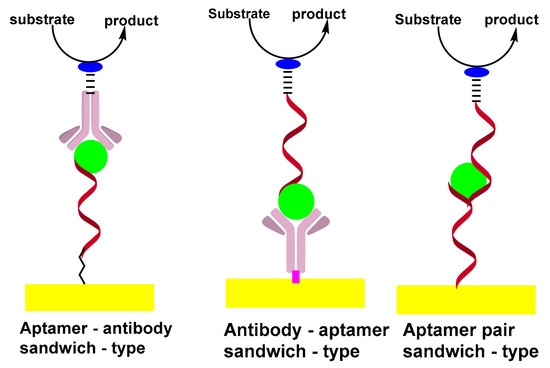
Scheme 2. Formation of sandwich-types structures of aptasensing assay.
2.2. Displacement-Type Electrochemical Aptasensors
In the displacement assay, the authentic target molecules compete with labeled target molecules for specific binding sites on the sensor surface. It was observed that labeled target molecules used as biocomponents in the displacement test exhibited less affinity than authentic targets towards the binding sites of aptamers[28]. This phenomenon was exploited by labeling the thrombin by peroxidase, thus modifying its structure and allowing it to be replaced by unmodified thrombin. In this format, the peroxidase-labeled thrombin was first captured by self-assembled monolayers (SAMs) of thiolated aptamer on the surface of the Au electrode. The sensor was then incubated with thrombin. The HRP confined to the electrode surface, was measured electrochemically by a H2O2 peroxidase catalyzed reaction[29]. Scheme 3 shows the displacement-type electrochemical aptasensor.
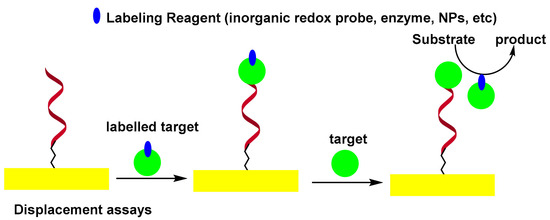
Scheme 3. Displacement-type electrochemical aptasensor.
2.3. Folding-Based Electrochemical Aptasensor
For decades, biomolecular recognition has prompted tremendous efforts to adapt biomolecules to the universal platform of molecular sensing. However, biosensors are almost completely unable to play their potential as a reagent-free real-time analysis device. The fundamental obstacle to the wider success of biosensors is that most biomolecules cannot generate easily measurable signals after binding to the target. For example, the antibody will not change its shape when it binds to the recognition target. Therefore, it has proven difficult to convert biomolecule binding events into measurable output signals, especially signals that are generated by the binding of many potentially interfering species in typical biological samples.
A potential solution to solve the problem is based on the binding of electrode-bound DNA probes to induce folding. In this sense, a series of electrochemical nucleic acid sensors using target-sensitive DNA structures with appropriate redox markers can monitor aptamer conformational changes by measuring electrochemical signals, which are directly dependent on the distance between the redox label and the electrode surface[30][31][32][33]. In an early work by Radi et al., a “Switch-on” aptasensor for the detection of thrombin was designed[30]. A thrombin binding aptamer with redox active Fc and thiol at the chain ends was self-assembled on the Au electrode by the S–Au bond. After thrombin interaction, the Fc-labeled aptamer changed its structure from random coil configuration of the aptamer to quadruplex, bringing the Fc label close to the electrode surface to facilitate the electron transfer between the Fc and the surface, as evidenced by the CV and EIS measurements. Scheme 4 illustrates the folding-based electrochemical aptasensor.
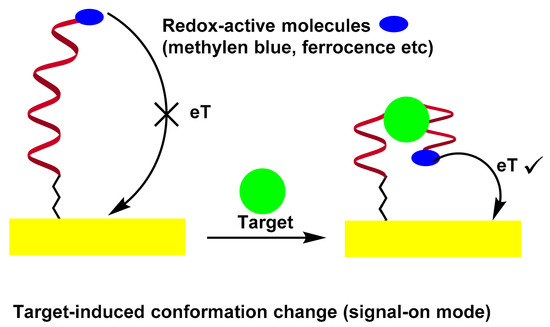
Scheme 4. Folding-Based Electrochemical Aptasensor.
Another electrochemical “Switch-off” aptasensor for the detection of thrombin was designed by Plaxco et al.[31]. A 32-base thrombin aptamer modified with thiol and MB at both ends was self-assembled on an Au electrode surface. When Tb was combined with the MB labeled aptamer, the MB marker was distant from the electrode surface and directly led to the weakening of the electrochemical signal.
3. Principles of Electrochemical Transduction Techniques
One of the biosensor components is the transducer, which converts the recognition event due to the interaction of the analyte with the recognition element into a measurable sign. The most important electroanalytical techniques are potentiometry, coulometry, voltammetry, amperometry, and electrochemical impedance spectroscopy (EIS). The electrochemical transducer is related to the reaction that involves electrons transfer. Therefore, electrochemical measurements can only be achieved with redox labels that can be attached to the target, recognition element, or soluble redox mediators, which diffuse onto the electrode surface to be reduced or oxidized by heterogeneous electron transfer. The redox probe includes redox enzymes, inorganic catalysts, or nanoparticles. The diffusion kinetics of the redox mediator to the electrode is influenced by the surface state of the transducer. Thus, the binding of the analyte affects the diffusion efficiency, electron transfer, and finally the detected signal. Other transduction approaches are connected to the intrinsic electroactivity of guanine-rich guanine aptamer on a graphite pencil electrode[34].
Nanoparticles can charge several units of electroactive species (ferrocene or thionine (Th)), and are thus used for signal amplification[35]. An efficient strategy is to load one or more enzymes (natural peroxidase and G-quadruplex/hemin (GQH) DNAzyme) as biocatalysts onto nanomaterials to improve the sensors’ sensitivity through enzymatic electrochemical processes; for example, a signal enhancement has been achieved by PCN-224-Pt nanoelectrocatalysts, natural HRP, and GQH HRP-mimicking DNAzymes catalyzing the oxidation of HQ with H2O2[36]. Natural redox enzymes could be replaced by catalytic nanomaterials as reporter molecules. In this respect, the nanoparticles CuO [37] and Pd–Au[38] show hydrogen peroxide reduction activity. Although they have the best stability against temperature and pH changes, their peroxidase activity is lower than that of the natural counterpart.
References
- Syed, M.A.; Pervaiz, S. Advances in Aptamers. Oligonucleotides 2010, 20, 215–224.
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822.
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510.
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9.
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug Deliv. Rev. 2006, 58, 671–685.
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242.
- Mascini, M. Aptamers and their applications. Anal. Bioanal. Chem. 2008, 390, 987–988.
- Ferreira, C.S.; Missailidis, S. Aptamer-based therapeutics and their potential in radiopharmaceutical design. Braz. Arch. Biol. Technol. 2007, 50, 63–76.
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650.
- Han, K.; Liang, Z.; Zhou, N. Design strategies for aptamer-based biosensors. Sensors 2010, 10, 4541–4557.
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical detection of protein using a double aptamer sandwich. Anal. Lett. 2004, 37, 2901–2909.
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172.
- Kara, P.; de la Escosura-Muñiz, A.; Maltez-da Costa, M.; Guix, M.; Ozsoz, M.; Merkoçi, A. Aptamers based electrochemical biosensor for protein detection using carbon nanotubes platforms. Biosens. Bioelectron. 2010, 26, 1715–1718.
- Lu, Y.; Zhu, N.; Yu, P.; Mao, L. Aptamer-based electrochemical sensors that are not based on the target binding-induced conformational change of aptamers. Analyst 2008, 133, 1256–1260.
- Yan, Z.; Han, Z.; Huang, H.; Shen, H.; Lu, X. Rational design of a thrombin electrochemical aptasensor by conjugating two DNA aptamers with G-quadruplex halves. Anal. Biochem. 2013, 442, 237–240.
- Zheng, J.; Lin, L.; Cheng, G.; Wang, A.; Tan, X.; He, P.; Fang, Y. Study on an electrochemical biosensor for thrombin recognition based on aptamers and nano particles. Sci. China Ser. B Chem. 2007, 50, 351–357.
- Xu, D.; Xu, D.; Yu, X.; Liu, Z.; He, W.; Ma, Z. Label-Free Electrochemical Detection for Aptamer-Based Array Electrodes. Anal. Chem. 2005, 77, 5107–5113.
- Numnuam, A.; Chumbimuni-Torres, K.Y.; Xiang, Y.; Bash, R.; Thavarungkul, P.; Kanatharana, P.; Pretsch, E.; Wang, J.; Bakker, E. Aptamer-Based Potentiometric Measurements of Proteins Using Ion-Selective Microelectrodes. Anal. Chem. 2008, 80, 707–712.
- Wang, X.; Zhou, J.; Yun, W.; Xiao, S.; Chang, Z.; He, P.; Fang, Y. Detection of thrombin using electrogenerated chemiluminescence based on Ru(bpy)32+-doped silica nanoparticle aptasensor via target protein-induced strand displacement. Anal. Chim. Acta 2007, 598, 242–248.
- Feng, K.; Sun, C.; Kang, Y.; Chen, J.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Label-free electrochemical detection of nanomolar adenosine based on target-induced aptamer displacement. Electrochem. Commun. 2008, 10, 531–535.
- Cho, E.J.; Lee, J.-W.; Ellington, A.D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264.
- Mir, M.; Vreeke, M.; Katakis, I. Different strategies to develop an electrochemical thrombin aptasensor. Electrochem. Commun. 2006, 8, 505–511.
- Seo, H.B.; Gu, M.B. Aptamer-based sandwich-type biosensors. J. Biol. Eng. 2017, 11, 11.
- Lee, S.J.; Park, J.-W.; Kim, I.-A.; Youn, B.-S.; Gu, M.B. Sensitive detection of adipokines for early diagnosis of type 2 diabetes using enzyme-linked antibody-aptamer sandwich (ELAAS) assays. Sens. Actuators B Chem. 2012, 168, 243–248.
- Shen, C.; Liu, S.; Li, X.; Yang, M. Electrochemical Detection of Circulating Tumor Cells Based on DNA Generated Electrochemical Current and Rolling Circle Amplification. Anal. Chem. 2019, 91, 11614–11619.
- Sun, D.; Lu, J.; Luo, Z.; Zhang, L.; Liu, P.; Chen, Z. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14.
- Miao, P.; Tang, Y. Gold Nanoparticles-Based Multipedal DNA Walker for Ratiometric Detection of Circulating Tumor Cell. Anal. Chem. 2019, 91, 15187–15192.
- Baldrich, E.; Acero, J.L.; Reekmans, G.; Laureyn, W.; O’Sullivan, C.K. Displacement enzyme linked aptamer assay. Anal. Chem. 2005, 77, 4774–4784.
- Mir, M.; Jenkins, A.T.A.; Katakis, I. Ultrasensitive detection based on an aptamer beacon electron transfer chain. Electrochem. Commun. 2008, 10, 1533–1536.
- Radi, A.E.; O’Sullivan, C.K. Aptamer conformational switch as sensitive electrochemical biosensor for potassium ion recognition. Chem. Commun. 2006, 3432–3434.
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991.
- Sánchez, J.L.A.; Baldrich, E.; Radi, A.E.G.; Dondapati, S.; Sánchez, P.L.; Katakis, I.; O’Sullivan, C.K. Electronic ‘off-on’ molecular switch for rapid detection of thrombin. Electroanalysis 2006, 18, 1957–1962.
- Zhang, Y.-L.; Huang, Y.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Electrochemical aptasensor based on proximity-dependent surface hybridization assay for single-step, reusable, sensitive protein detection. J. Am. Chem. Soc. 2007, 129, 15448–15449.
- Kawde, A.-N.; Rodriguez, M.C.; Lee, T.M.; Wang, J. Label-free bioelectronic detection of aptamer–protein interactions. Electrochem. Commun. 2005, 7, 537–540.
- Dou, B.; Xu, L.; Jiang, B.; Yuan, R.; Xiang, Y. Aptamer-Functionalized and Gold Nanoparticle Array-Decorated Magnetic Graphene Nanosheets Enable Multiplexed and Sensitive Electrochemical Detection of Rare Circulating Tumor Cells in Whole Blood. Anal. Chem. 2019, 91, 10792–10799.
- Ou, D.; Sun, D.; Liang, Z.; Chen, B.; Lin, X.; Chen, Z. A novel cytosensor for capture, detection and release of breast cancer cells based on metal organic framework PCN-224 and DNA tetrahedron linked dual-aptamer. Sens. Actuators B Chem. 2019, 285, 398–404.
- Tian, L.; Qi, J.; Qian, K.; Oderinde, O.; Liu, Q.; Yao, C.; Song, W.; Wang, Y. Copper (II) oxide nanozyme based electrochemical cytosensor for high sensitive detection of circulating tumor cells in breast cancer. J. Electroanal. Chem. 2018, 812, 1–9.
- Liu, J.-X.; Bao, N.; Luo, X.; Ding, S.-N. Nonenzymatic Amperometric Aptamer Cytosensor for Ultrasensitive Detection of Circulating Tumor Cells and Dynamic Evaluation of Cell Surface N-Glycan Expression. ACS Omega 2018, 3, 8595–8604.





