
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mariarosaria Conte | + 1460 word(s) | 1460 | 2021-01-18 09:16:19 | | | |
| 2 | Catherine Yang | Meta information modification | 1460 | 2021-01-19 10:28:56 | | | | |
| 3 | Catherine Yang | Meta information modification | 1460 | 2021-01-26 04:39:49 | | |
Video Upload Options
Sessile organisms such as seaweeds, corals, and sponges continuously adapt to both abiotic and biotic components of the ecosystem. This extremely complex and dynamic process often results in different forms of competition to ensure the maintenance of an ecological niche suitable for survival. A high percentage of marine species have evolved to synthesize biologically active molecules, termed secondary metabolites, as a defense mechanism against the external environment. These natural products and their derivatives may play modulatory roles in the epigenome and in disease-associated epigenetic machinery. Epigenetic modifications also represent a form of adaptation to the environment and confer a competitive advantage to marine species by mediating the production of complex chemical molecules with potential clinical implications. Bioactive compounds are able to interfere with epigenetic targets by regulating key transcriptional factors involved in the hallmarks of cancer through orchestrated molecular mechanisms, which also establish signaling interactions of the tumor microenvironment crucial to cancer phenotypes.
1. Introduction
Marine habitats are an extraordinary source of new and structurally complex bioactive metabolites naturally produced by different organisms and characterized by unique functions with marked biological activities. These features can be attributed to extreme environmental conditions such as lack of light, high pressure, ionic concentration, pH and temperature changes, scarcity of nutrients, and restricted living spaces [1]. The high concentration of coexisting organisms in a limited area also makes them very competitive and complex, resulting in the development of adaptations and behaviors aimed at safeguarding the species, such as the adoption of chemical strategies exploiting the wealth of bioactive molecules produced by the secondary metabolism [2][3]. Marine-derived metabolites originate from different signal transduction pathways activated as a consequence of epigenome changes in the organisms that produce them. Phenotypic/genotypic alterations of marine organisms are characterized by an intricate network of interactions that influence each other. Such interplay is further complicated by epigenetic modifications, which can trigger adaptive biochemical processes in the species [4]. The marine environment (characterized by biotic and abiotic factors), in turn, plays an essentially selective role in intrinsically changing organisms, exerting an inductive function on epigenetic, genetic, and phenotypic changes with transgenerational effects on the species [5][6] (Figure 1). Since the reprogramming of epigenetic states can be induced by environmental exposures in the marine habitat, secondary metabolites produced by a large number of organisms might represent good candidates as novel natural molecules with potential pharmacological activity for cancer treatment [7]. Cutting-edge chromatographic isolation and purification techniques, pharmacological screening methods, and numerous spectroscopic approaches for structural investigation such as mass spectroscopy and nuclear magnetic resonance (NMR) were used to isolate and characterize several new marine-derived compounds [8][9][10], some of which have potential anticancer activities. The chemical composition of isolated molecules has a major impact on both the epigenome of the organism [11] and any potential epigenetic effects produced, reflecting a complex and interconnected machinery of information exchange. In this review, we describe the therapeutic potential of marine-derived secondary metabolites and their synthetic derivatives in cancer, focusing on their importance as epigenetic modulators generating posttranscriptional, inductive (produced by the metabolism of the organism), and induced (produced by alterations in marine environment) modifications. We also discuss the challenges involved in discovering new natural and synthetic marine bio-compounds with anticancer activity in light of the enormous variability that characterizes the organisms themselves and the environment that surrounds them. This review also highlights sustainable use of marine resources as producers of high yields of value added bio-molecules for pharmaceutical field towards a more sustainability of economic growth in terms of development, research and transmissibility of marine technology in terms of development, research, and transmissibility of marine technology.
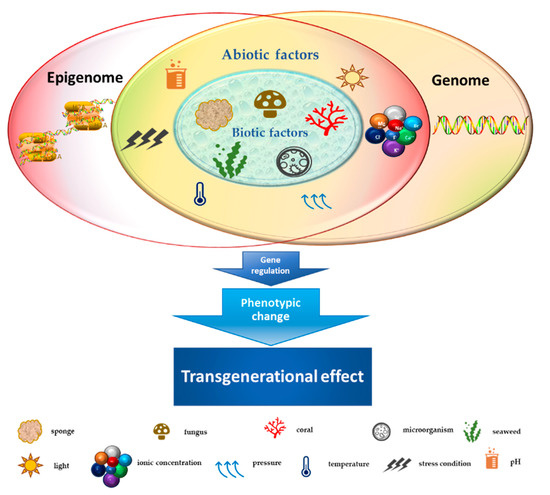
Figure 1. Schematic representation of the crosstalk between biotic/abiotic factors and transgenerational genetic/epigenetic effects.
2. Anticancer Activities of Marine-Derived Secondary Metabolites with Inductive and Induced Epigenetic Modifications
Epigenetics belongs to a branch of genetics based on the concomitance of complex biomolecular mechanisms, which coordinate genetic information in the nucleus, culminating in control of gene expression [12], which in turn propagates in subsequent generations. All this information is further conditioned by perturbations from the external environment. Epigenetic alterations in gene activity are mitotically stable in the absence of changes in DNA sequence [13]. Generally, mechanisms of environmental perception act through alterations in chemical tags that normally exist in the genome. In cancer, “epigenetic markers” [14] serve as a sort of barcode of DNA function, indicating whether genes are active or silent. The alteration and reprogramming of epi-signals can lead to changes in gene expression and also directly influence transcriptional regulator function, with downstream effects on the way cells and tissues work. In addition to genetic alterations, a hallmark of various types of cancer, epigenetic dysregulations affecting DNA methylation, histone modifications, and microRNAs introduce another layer of complexity, contributing to tumor progression and changes in the phenotypic state. These epi-alterations are further regulated by so-called chromatin writers, readers, and erasers, which constitute specialized protein machinery able to modulate and reversibly influence the epigenome. DNA methyltransferases (DNMTs), histone acetyltransferases (HATs), histone methyltransferases (HMTs), and lysine/arginine methyltransferases (KMTs/RMTs) are all writers, due to their ability to add a modification on DNA and histones; readers, able to “read” and thus interpret covalent modifications include: bromodomains, specific for acetylation site recognition; chromodomains, recognized by methyl-readers; methyl-lysine readers such as ATRX-DNMT3-DNMT3L (ADD), ankyrin, bromo-adjacent homology, chromobarrel, WD40, and zinc finger CW domains, as well as double chromodomain, and tandem Tudor domain. In addition, plant homeodomains bind to methylated histone H3, while the PWWP domain can bind DNA and methylated histones. Erasers include TET proteins, which remove modifications from DNA and histones, as well as histone demethylases (HDMs), histone deacetylases (HDACs), protein phosphatases, and deubiquitinating enzymes, which remove methyl, acetyl, phosphate, and ubiquitin groups from histones and other proteins, respectively [15][16][17]. Since modern medical approaches are based on the personalization of human healthcare, bioactive molecules with epigenetic activity isolated from marine sources represent a valid alternative to conventional therapies for use in extensive preclinical assessments and the advanced phases of clinical studies. Furthermore, given the resistance developed by some pathogens to pharmacological treatments and the inefficacy of traditional chemotherapies, efforts are being made to identify more biologically active and effective molecules [18]. In marine ecosystems, sessile organisms are much more susceptible to changes in the external environment [19] and adopt complex survival strategies. Moreover, the set of biotic and abiotic components in these organisms are extremely predominant, determining the production of secondary metabolites with almost unique chemical-physical characteristics. A further level of complexity is added by the intricate relationship between secondary metabolites and epigenetic functions, which in turn contribute to the development of defense mechanisms by the species that are transmitted across generations [20]. Secondary metabolites can harbor several beneficial properties for human health such as antioxidant, antibacterial, antivirus, anticoagulant, antidiabetic, anti-inflammatory, antihypertensive, and antitumor activities [21]. Furthermore, their natural biological functions are strongly influenced by the surrounding environment, including conditions of climatic stress or attack by predators. Computational programs using knowledge-based algorithms or sequence-based prediction [22] have identified genes responsible for the production of these natural products, but only for some species. These genes are usually located in specific biosynthetic gene clusters (BGCs) in the genome [23] that contain the required enzymes responsible for synthesis of secondary metabolites and regulatory structures. Considering the enormous genetic and epigenetic variability among marine species, it is not always possible to predict their BGCs and therefore their association with the production of secondary metabolites. For example, under certain conditions chromatin remodeling factors can switch on or switch off specific genes continuously over time. Cutting-edge technologies such as those involved in triggering the activation of silent BGCs, which include changes in growth conditions (e.g., temperature and pH) or genetic engineering-based approaches [24][25] are emerging to better study the interaction between the production of metabolites and the genes that produce them. One of the goals of anticancer research is to extract and select biomolecules from these organisms in order to exploit their properties and generate synthetic analogues (Figure 2). To date, the U.S. Food and Drug Administration (FDA) has approved several marine-derived therapeutic compounds such as cytarabine, vidarabine, ziconotide, omega-3 acid ethyl esters, eribulin mesylate, brentuximab vedotin, and iota-carrageenan [26][27][28][29][30][31][32] (Figure 3) and further studies aimed at characterizing and developing new drugs are ongoing. The following section describes the epigenetic role of well-known marine-derived secondary metabolites, classified according to their biosynthetic pathways and subdivided into three major families: phenolic compounds, cyclic peptides, and alkaloids. Their mechanism of action as potential epigenetic bio-compounds for the treatment of different type of cancers is also discussed.
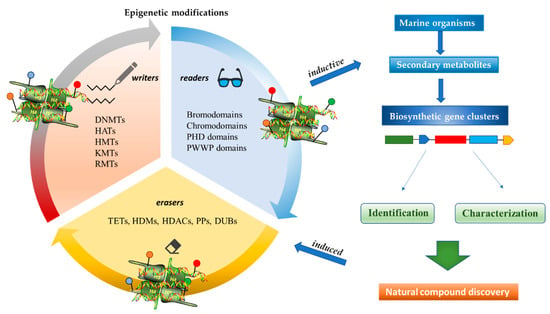
Figure 2. Inductive and induced epigenetic modifications by secondary metabolites produced by marine organisms. Epigenetic writers, readers, and erasers regulate production of secondary metabolites, which in turn induce epi-modifications.
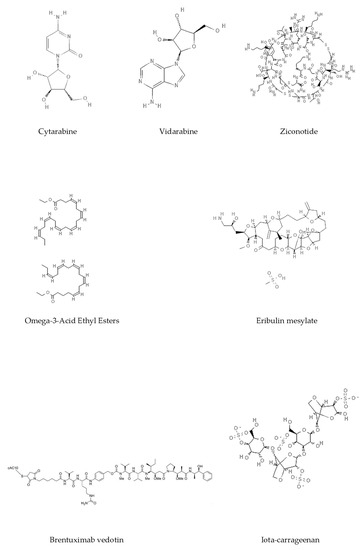
Figure 3. Chemical structures of marine-derived therapeutic compounds approved by the U.S. Food and Drug Administration (FDA).
References
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms 2017, 5, 25.
- Firn, R.D.; Jones, C.G. The evolution of secondary metabolism-a unifying model. Mol. Microbiol. 2000, 37, 989–994.
- Giordano, D.; Coppola, D.; Russo, R.; Denaro, R.; Giuliano, L.; Lauro, F.M.; di Prisco, G.; Verde, C. Marine Microbial Secondary Metabolites: Pathways, Evolution and Physiological Roles. Adv. Microb. Physiol. 2015, 66, 357–428.
- Carneiro, V.C.; Lyko, F. Rapid Epigenetic Adaptation in Animals and Its Role in Invasiveness. Integr. Comp. Biol. 2020, 60, 267–274.
- Mirbahai, L.; Chipman, J.K. Epigenetic memory of environmental organisms: A reflection of lifetime stressor exposures. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 764–765, 10–17.
- Jeremias, G.; Barbosa, J.; Marques, S.M.; Asselman, J.; Goncalves, F.J.M.; Pereira, J.L. Synthesizing the role of epigenetics in the response and adaptation of species to climate change in freshwater ecosystems. Mol. Ecol. 2018, 27, 2790–2806.
- Seca, A.M.L.; Pinto, D. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263.
- Kiuru, P.; D’Auria, M.V.; Muller, C.D.; Tammela, P.; Vuorela, H.; Yli-Kauhaluoma, J. Exploring marine resources for bioactive compounds. Planta Med. 2014, 80, 1234–1246.
- Lindequist, U. Marine-Derived Pharmaceuticals-Challenges and Opportunities. Biomol. Ther. 2016, 24, 561–571.
- Sun, W.; Wu, W.; Liu, X.; Zaleta-Pinet, D.A.; Clark, B.R. Bioactive Compounds Isolated from Marine-Derived Microbes in China: 2009–2018. Mar. Drugs 2019, 17, 339.
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167.
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505.
- D’Urso, A.; Brickner, J.H. Mechanisms of epigenetic memory. Trends Genet. 2014, 30, 230–236.
- Thomas, M.L.; Marcato, P. Epigenetic Modifications as Biomarkers of Tumor Development, Therapy Response, and Recurrence across the Cancer Care Continuum. Cancers 2018, 10, 101.
- Yang, A.Y.; Kim, H.; Li, W.; Kong, A.N. Natural compound-derived epigenetic regulators targeting epigenetic readers, writers and erasers. Curr. Top. Med. Chem. 2016, 16, 697–713.
- Torres, I.O.; Fujimori, D.G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015, 35, 68–75.
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24.
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614.
- Bosch, T.C.; Adamska, M.; Augustin, R.; Domazet-Loso, T.; Foret, S.; Fraune, S.; Funayama, N.; Grasis, J.; Hamada, M.; Hatta, M.; et al. How do environmental factors influence life cycles and development? An experimental framework for early-diverging metazoans. Bioessays 2014, 36, 1185–1194.
- Dias, B.G.; Maddox, S.; Klengel, T.; Ressler, K.J. Epigenetic mechanisms underlying learning and the inheritance of learned behaviors. Trends Neurosci. 2015, 38, 96–107.
- Seca, A.M.L.; Pinto, D. Biological Potential and Medical Use of Secondary Metabolites. Medicines 2019, 6, 66.
- Vetrivel, I.; Mahajan, S.; Tyagi, M.; Hoffmann, L.; Sanejouand, Y.H.; Srinivasan, N.; de Brevern, A.G.; Cadet, F.; Offmann, B. Knowledge-based prediction of protein backbone conformation using a structural alphabet. PLoS ONE 2017, 12, e0186215.
- Khalifa, S.A.M.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.F.; Moustafa, M.S.; Abd El-Wahed, A.; Al-Mousawi, S.M.; Musharraf, S.G.; et al. Marine Natural Products: A Source of Novel Anticancer Drugs. Mar. Drugs 2019, 17, 491.
- Lin, D.; Xiao, M.; Zhao, J.; Li, Z.; Xing, B.; Li, X.; Kong, M.; Li, L.; Zhang, Q.; Liu, Y.; et al. An Overview of Plant Phenolic Compounds and Their Importance in Human Nutrition and Management of Type 2 Diabetes. Molecules 2016, 21, 1374.
- Baral, B.; Akhgari, A.; Metsa-Ketela, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178.
- Tomizawa, D.; Tanaka, S.; Hasegawa, D.; Iwamoto, S.; Hiramatsu, H.; Kiyokawa, N.; Miyachi, H.; Horibe, K.; Saito, A.M.; Taga, T.; et al. Evaluation of high-dose cytarabine in induction therapy for children with de novo acute myeloid leukemia: A study protocol of the Japan Children’s Cancer Group Multi-Center Seamless Phase II-III Randomized Trial (JPLSG AML-12). Jpn. J. Clin. Oncol. 2018, 48, 587–593.
- Tamborini, L.; Previtali, C.; Annunziata, F.; Bavaro, T.; Terreni, M.; Calleri, E.; Rinaldi, F.; Pinto, A.; Speranza, G.; Ubiali, D.; et al. An Enzymatic Flow-Based Preparative Route to Vidarabine. Molecules 2020, 25, 1223.
- McDowell, G.C., 2nd; Pope, J.E. Intrathecal Ziconotide: Dosing and Administration Strategies in Patients with Refractory Chronic Pain. Neuromodulation 2016, 19, 522–532.
- Czyz, K.; Sokola-Wysoczanska, E.; Bodkowski, R.; Cholewinska, P.; Wyrostek, A. Dietary Omega-3 Source Effect on the Fatty Acid Profile of Intramuscular and Perimuscular Fat-Preliminary Study on a Rat Model. Nutrients 2020, 12, 3382.
- Nakano, K.; Hayakawa, K.; Funauchi, Y.; Tanizawa, T.; Ae, K.; Matsumoto, S.; Tomomatsu, J.; Ono, M.; Taira, S.; Nishizawa, M.; et al. Differences in the efficacy and safety of eribulin in patients with soft tissue sarcoma by histological subtype and treatment line. Mol. Clin. Oncol. 2020, 14, 13.
- Oberic, L.; Delzor, F.; Protin, C.; Perriat, S.; Laurent, C.; Grand, A.; Canonge, J.M.; Borel, C.; Gauthier, M.; Ysebaert, L.; et al. Brentuximab vedotin in real life, a seven year experience in patients with refractory/relapsed CD30+ T cell lymphoma. J. Oncol. Pharm. Pract. 2020, 1078155220968615.
- Eccles, R.; Winther, B.; Johnston, S.L.; Robinson, P.; Trampisch, M.; Koelsch, S. Efficacy and safety of iota-carrageenan nasal spray versus placebo in early treatment of the common cold in adults: The ICICC trial. Respir. Res. 2015, 16, 121.




