| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Paolo Grumati | + 1218 word(s) | 1218 | 2020-11-10 09:53:11 | | | |
| 2 | Karina Chen | -19 word(s) | 1199 | 2021-01-19 10:22:42 | | |
Video Upload Options
The aim of the review manuscript is to provide an over-view of the close relation between ubiquitin and the selective process of autophagy. We described how ubiquitin determine the selectivity towards different cellular component and how it may influence autophagy receptors activity. Precisely, we discussed the role of the ubiquitin signal in each type of selective autophagy.
1. Introduction
The cellular life cycle is complex, having to contend with ever-changing and at times competing internal and environmental demands, can be stressful. Fail-safe degradation mechanisms are therefore required for the effective disposal of potentially toxic and harmful components and their recycling into building blocks needed for biosynthesis. These degradation systems are vital for the survival and continuity of both long-lived and dividing cells. Several cellular degradation processes have evolved to fill this need and their importance is illustrated through their conservation across evolution, and the pathology that ensues with their perturbance [1][2].
The ubiquitin-proteasome and autophagy-lysosome are the two major cellular degradation systems found in eukaryotic cells and organisms. These processes have remained conserved among species and failure of either one can result in the accumulation of toxic or damaged proteins and organelles, culminating in a number of severe pathologies including cancer, failure to thrive, degenerative diseases, and premature death [1][2]. The ubiquitin-proteasome pathway mainly relies on the 26S proteasome for the final degradation of its substrates [3]. Considered to be the more selective of the two degradation systems, proteasomal substrates are largely composed of individual proteins, requiring large complexes to be disassembled before degradation can take place [4]. The autophagy-lysosome pathway utilizes double membraned vesicles, termed autophagosomes, for the encapsulation and delivery of components to the lysosome for breakdown [5]. Autophagy substrates tend to be larger, including protein aggregates, organelles- either in their entirety or select portions, and invading pathogens. Although autophagy was historically considered to be a bulk degradation pathway, it is now universally accepted that it can be quite selective, with a wide range of substrates under its jurisdiction. Despite the two degradation mechanisms being fairly distinct, they both appear to utilize ubiquitin modification for substrate recognition [6]. It is intriguing that two seemingly independent degradation pathways, which have evolved largely different components and substrates, converge on the same PTM for cargo recognition. This suggests some crosstalk and redundancy between these pathways, with ubiquitin acting as a universal degradation signal.
2. Autophagy
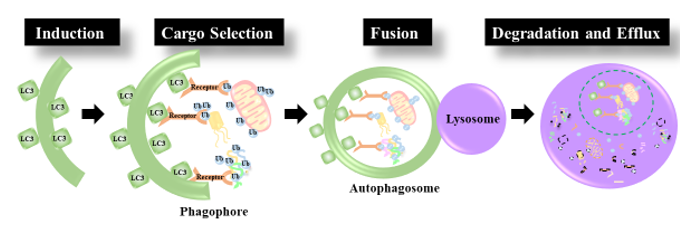
Macroautophagy, herein on termed Autophagy, is a cellular recycling mechanism responsible for cellular housekeeping and turnover during steady-state conditions and nutrient scavenging during starvation. This process involves the formation of double membraned vesicles, known as autophagosomes, around a substrate or a cytoplasmic domain, and the subsequent delivery of these vesicles to the lysosome for proteolytic degradation (Figure 1). Autophagy depends on core autophagy proteins which contribute to the initiation of autophagosome formation, the lipidation of the autophagosomal membrane protein Atg8/LC3/GABARAP, the fusion of the autophagosome to the lysosome, and finally, the degradation of the autophagosome with its cargo within the lysosomal lumen [7]. In 2016 Yoshinori Ohsumi received the Nobel prize in Physiology and Medicine for his discovery of the mechanisms of autophagy in yeast. Ohsumi systematically uncovered autophagy-related genes, classified as ATGs, and his discoveries resulted in an autophagy renaissance.
Figure 1. Autophagy Induction: first a pre-autophagosome (phagophore) is formed by the lipidation of LC3. Cargo Selection: Ubiquitinated cargo is tagged for degradation by autophagy receptors equipped with an LC3 binding motif and a ubiquitin binding domain. The Autophagosome is then elongated around the cargo to be degraded. Fusion: the mature autophagosome fuses with the lysosome. Degradation and efflux: The contents of the autophagosome are degraded by proteolytic enzymes within the lysosomal lumen and nutrients are released.
Once considered an indiscriminate bulk degradation process, autophagy is now well recognized for its selectivity. With the aid of various receptors and signaling molecules, autophagy can discern between healthy cellular components and toxic organelles, proteins, as well as invading organisms. Indeed, constitutive autophagy can be extremely selective and is responsible for the removal of cellular components, like misfolded protein aggregates or exhausted organelles, to ensure a proper physiological turnover [8]. Moreover, autophagy induced by a specific trigger can also be very selective; for instance, xenophagy induced by a pathogen infection triggers a selective autophagic response against the invading organism. Likewise, when mitochondria are chemically depolarized, mitophagy ensues and targets strictly depolarized mitochondria [9][10][11]. Contrarily, autophagy induced by severe nutrient depletion may not be overly selective, as its sole goal is nutrient liberation under conditions of cellular stress. With this in mind, various types of substrates are eliminated through selective autophagy and this number is continuously on the rise. During selective autophagy, dysfunctional or obsolete proteins and organelles are targeted for degradation by various receptors, which, in turn, entice the arrival of the autophagosome. The selectivity in this process is largely mediated by PTMs such as phosphorylation and ubiquitination, with UFMylation, ISGylation, and SUMOylation recently arising as potential PTMs, similar to Ubiquitination, that modulate autophagy [12].
3. Selective autophagy
The benefits of selective autophagy are vast, as it allows for the surgical removal of targeted substrates. For instance, the removal of mitochondria during hypoxia, organelle damage, or even when the organelle becomes obsolete, such as during erythrocyte maturation, all occur selectively through mitophagy [13][14][15]. In bulk autophagy, the ubiquitination of autophagic components often acts as a regulatory signal. However, when it comes to selective autophagy ubiquitination also acts as a signal for cargo recognition, process initiation, and rate of autophagosome formation.
Generally, during selective autophagy, protein aggregates, damaged organelles or portions thereof are identified and tagged for degradation by E3 Ub-ligases. Once cargos are tagged, ubiquitin acts as an “eat me” signal for autophagy receptors, which further flag targets for degradation. Autophagy receptors are also implicated in segregating and coalescing materials destined for autophagic degradation, effectively preparing them for the arrival of the phagophore. Indeed, autophagy receptors harbor a UBL domain to sense Ub molecules, and a LIR domain to bind the LC3s/GABARAPs, which are present on autophagosomal membranes. By binding to mATG8s, cargo receptors promote the recruitment of autophagosomal membranes around the Ub-tagged materials, eventually encapsulating them completely. Mature autophagosomes are then delivered to the lysosome for degradation [16][17].
Considering that potentially all cytosolic components could be selectively degraded via lysosomes, it is plausible that selective autophagy plays a role in a number of diverse physiological processes and their associated diseases. The ability to specifically target the elimination of desired cellular constituents presents great therapeutic potential for the treatment of many diseases characterized by the accumulation of toxic materials including neurodegenerative diseases like Huntington’s and Alzheimer’s, lysosomal storage diseases, and mitochondrial DNA disorders [18][19]. Moreover, selective autophagy has also been implicated in chronic obstructive pulmonary disease (COPD) [20] as well as other pulmonary disorders [21]. Of note, one of the most well-studied functions of selective autophagy in human health is its role in infections [22]. Once bacteria invade the host cell, they are immediately labeled with ubiquitin chains and selectively degraded via xenophagy [23]. The removal of viral pathogens by selective autophagy is also termed virophagy [24][25]. Of note, host cells counteract some types of viral infections, like flaviviruses and Ebola virus, by directly eliminating their replicative niche in the endoplasmic reticulum, thus activating another form of selective autophagy named ER-phagy [26][27].
References
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta–Mol. Cell Res. 2014, 1843, 13–25.
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995.
- Kleiger, G.; Mayor, T. Perilous journey: A tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014, 24, 352–359.
- Papadopoulos, C.; Kirchner, P.; Bug, M.; Grum, D.; Koerver, L.; Schulze, N.; Poehler, R.; Dressler, A.; Fengler, S.; Arhzaouy, K.; et al. VCP/p97 cooperates with YOD 1, UBXD 1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017, 36, 135–150.
- Piccirillo, R.; Goldberg, A.L. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 2012, 31, 3334–3350.
- Kraft, C.; Peter, M.; Hofmann, K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010, 12, 836–841.
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12.
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42.
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14.
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285.
- Georgakopoulos, N.D.; Wells, G.; Campanella, M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 2017, 13, 136–146.
- Nakka, V.P.; Mohammed, A.Q. A Critical Role for ISGylation, Ubiquitination and, SUMOylation in Brain Damage: Implications for Neuroprotection. Neurochem. Res. 2020, 45, 1975–1985.
- Bellot, G.; Garcia-Medina, R.; Gounon, P.; Chiche, J.; Roux, D.; Pouysségur, J.; Mazure, N.M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 2009, 29, 2570–2581.
- Jin, S.M.; Youle, R.J. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 2013, 9, 1750–1757.
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010, 11, 45–51.
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014, 16, 495–501.
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30.
- Scrivo, A.; Bourdenx, M.; Pampliega, O.; Cuervo, A.M. Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol. 2018, 17, 802–815.
- Dombi, E.; Mortiboys, H.; Poulton, J. Modulating Mitophagy in Mitochondrial Disease. Curr. Med. Chem. 2017, 25, 5597–5612.
- Mizumura, K.; Cloonan, S.M.; Nakahira, K.; Bhashyam, A.R.; Cervo, M.; Kitada, T.; Glass, K.; Owen, C.A.; Mahmood, A.; Washko, G.R.; et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Investig. 2014, 124, 3987–4003.
- Mizumura, K.; Choi, A.M.K.; Ryter, S.W. Emerging role of selective autophagy in human diseases. Front. Pharmacol. 2014, 5, 244.
- Reggio, A.; Buonomo, V.; Grumati, P. Eating the unknown: Xenophagy and ER-phagy are cytoprotective defenses against pathogens. Exp. Cell Res. 2020, 396, 112276.
- Sharma, V.; Verma, S.; Seranova, E.; Sarkar, S.; Kumar, D. Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 2018, 6, 147.
- Dong, X.; Levine, B. Autophagy and viruses: Adversaries or allies? J. Innate Immun. 2013, 5, 480–493.
- Mijaljica, D.; Klionsky, D.J. Autophagy/virophagy: A “disposal strategy” to combat COVID-19. Autophagy 2020, 1-2, doi: 10.1080/15548627.2020.1782022.
- Chiramel, A.I.; Dougherty, J.D.; Nair, V.; Robertson, S.J.; Best, S.M. FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga. J. Infect. Dis. 2016, 214, S319–S325.
- Lennemann, N.J.; Coyne, C.B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 2017, 13, 322–332.