| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Paweł Krzyżek | + 1068 word(s) | 1068 | 2020-12-29 07:59:42 | | | |
| 2 | Paweł Krzyżek | Meta information modification | 1068 | 2021-01-14 15:14:06 | | | | |
| 3 | Paweł Krzyżek | Meta information modification | 1068 | 2021-01-14 15:16:24 | | |
Video Upload Options
Microbial biofilm is defined as a structured consortium of cells immersed in a self-produced matrix. It is worth mentioning, however, that the biofilm may also include host components, e.g., fibrin, antibodies, platelets, or leukocytes. Biofilms can be attached to an abiotic or biotic surface, but they can also constitute a mobile, non-adhered structure floating in culture broth or body fluids.
1. Introduction
H. pylori has the ability to reside in the stomach for many years, often for a lifetime [1]. Although the presence of this bacterium may be associated with a number of gastrointestinal diseases, including gastric ulcers and cancers [2][3][4], more and more often the protective role against developing extra-gastric diseases, such as allergy, asthma, inflammatory bowel disease, or multiple sclerosis, is indicated [5][6][7]. Survival of H. pylori in an unfavorable, gastric niche is enabled by an intensive urease secretion, a spiral shape, presence of numerous adhesins, and the production of cytotoxic proteins, e.g., vacuolating cytotoxin A (VacA) and cytotoxin-associated gene A (CagA) [8]. It is increasingly recognized, however, that the adaptive mechanisms of H. pylori in the stomach may also be linked to the ability of this pathogen to form biofilms [9].
2. Transcriptomic and Proteomic Analysis of Biofilm H. pylori Forms
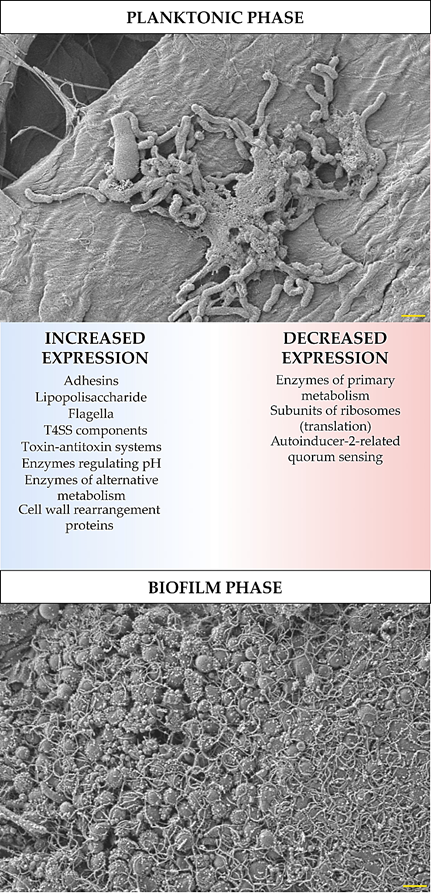
The development of biofilm structure and the transition from planktonic to biofilm phase are influenced by changes in the expression and translation of H. pylori genes. These changes were determined experimentally using ‘omics’ techniques (transcriptomics, proteomics, and metabolomics), and despite some differences between the results obtained by independent research teams, it is still possible to identify the most important features of the biofilm forms of this bacterium. In these studies, an increased expression has been demonstrated for: global pleiotropic regulators [10][11][12][13], adhesins (especially outer membrane proteins from the Hop and Hom family) [10][11][14][15], lipopolysaccharide (LPS) [10][11][14], efflux pumps [11][16][17][18][19][20], flagella proteins [10][21][14], components of T4SS systems [10][11][21][14], enzymes regulating pH (e.g., urease or arginase) [21][22] and responsible for obtaining alternative energy sources (e.g., hydrogenase) [11][21], proteins related to the cell wall rearrangement [11][21], and proteins of the toxin-antitoxin system [23] (Figure 1). Covering the components for which a decrease in the expression in biofilm H. pylori forms has been shown, these were factors involved in metabolism [10][11][24], translation [10][11], and quorum sensing related to the autoinducer-2 (AI-2) activity [11][25][26][27][28] (Figure 1). The influence of the transition of H. pylori into the biofilm form on the amount of antioxidant proteins is difficult to determine, because Yang et al. [22] and Shao et al. [21] showed their increase, while Hathroubi et al. [11] showed an opposite situation. It is possible that this difference results from the type of analyzed biofilm. Yang et al. [22] and Shao et al. [21] studied biofilm cells that were exposed to oxidative stress (air-liquid biofilms or these formed as microaggregates in the medium), while the one studied by Hathroubi et al. [11] was a sedentary biofilm produced at the bottom of wells, where the oxygen concentration is significantly lower.
Figure 1. Diagram showing transcriptomic/proteomic changes in H. pylori cells during the transition from planktonic to biofilm phase. The scale bar shows 1 µm.
Among the proteins mentioned above, it is worth paying attention to the production of global regulators of H. pylori physiology, because they will influence the expression of other genes under their control. In general, negative regulators bind to the promoter, reducing the attachment of RNA polymerase and lowering the expression of specific genes. In the case of positive regulators, binding is observed in the promoter's upstream region, which determines the recruitment of polymerase and the increase in gene expression [29][30].
Shao et al. [21] showed that aconitase production increases in the biofilm phase. Aconitase acts as a pleiotropic regulator supervising antioxidant functions, motility and production of flagella, as well as the activity of some enzymes (urease and hydrogenase) [31]. It was established that the high activity of H. pylori hydrogenase is a feature of strains with a high carcinogenic potential [32]. Through hydrogenase, this bacterium can use molecular hydrogen (H2) to switch to the chemolithoautotrophic mode and use the energy from this process to bind CO2 [33]. Hydrogenase is also crucial for the functioning of the T4SS systems, both Cag-T4SS (involved in cytotoxicity and oncogenicity) and ComB-T4SS (involved in DNA transformation) [32]. It is worth noting that the ComB system is inactive at acidic pH and is stimulated in the environment with pH > 6.5, indicating that the transformation process takes place in close proximity to the gastric mucosa [34] and explaining the increase in urease production during biofilm formation (when microbial transformation is strongly intensified) [22].
The increased expression of H. pylori regulators during the transition to the biofilm phase, including HspR, HrcA, CrdR, and RsfS, was also demonstrated by the team of Hathroubi et al. [10][11]. HspR and HrcA have a positive effect on the production of flagella and selected adhesins, while negative on the chaperone proteins (GroES, GroEL, and DnaK) [35][36]. CrdR responds in H. pylori to the nitrosic stress (exposure to NO) [12]. It was experimentally found that flagellar components, iron transporter (fecA, HP0807) and efflux pump (glnP, HP1169) were among the genes with the highest induction controlled by CrdR [12]. For RsfS, participation in the inhibition of large and small subunits of ribosomes has been shown and, as a consequence, a reduction of protein synthesis (one of the most energy-consuming physiological processes) [37]. This mechanism seems to explain the observations of Hathroubi et al., who noticed a decrease in the expression of translation genes during the transition of H. pylori to the biofilm phase [10][11].
Apart from global regulators having a positive effect on the production of biofilm, it is also worth paying attention to the existence of the ArsRS regulator, which is a two-component system negatively associated with the production of this structure [38][15]. For ArsRS, the pH-dependent effect is indicated, during which exposure to the acidic pH of gastric juice inhibits the expression of genes related to biofilm formation, while in an environment with a neutral pH the effect of this system is inhibited [38]. The ∆arsS mutants showed a higher degree of adhesion to the surface and more intensive biofilm production (twice after 1–2 days and four times after 3 days of incubation than the wild-type strain) [38][15]. Moreover, in these mutants, an increase in the expression of genes encoding outer membrane proteins was noticed; these included alpB (omp21/homB), sabA (omp17/hopP), labA (omp2/hopD), and hopZ (omp1) [15][39].
The presented data show that global regulators can significantly influence the physiology of H. pylori and determine the transition from planktonic to biofilm form and vice versa.
References
- Kwang-Ho Rhee; Jin-Sik Park; Myung-Je Cho; Helicobacter pylori: Bacterial Strategy for Incipient Stage and Persistent Colonization in Human Gastric Niches. Yonsei Medical Journal 2014, 55, 1453-1466, 10.3349/ymj.2014.55.6.1453.
- Milica Denic; Eliette Touati; Hilde De Reuse; Review: Pathogenesis of Helicobacter pylori Infection.. Helicobacter 2020, 25, e12736, doi.org/10.1111/hel.12736.
- Yoshio Yamaoka; David Y Graham; Helicobacter pylori virulence and cancer pathogenesis. Future Oncology 2014, 10, 1487-1500, 10.2217/fon.14.29.
- Christian Schulz; Juozas Kupčinskas; Review - Helicobacter pylori and non-malignant upper gastro-intestinal diseases. Helicobacter 2020, 25, e12738, 10.1111/hel.12738.
- Karen Robinson; Helicobacter pylori-Mediated Protection against Extra-Gastric Immune and Inflammatory Disorders: The Evidence and Controversies. Diseases 2015, 3, 34-55, 10.3390/diseases3020034.
- Rinaldo Pellicano; Gianluca Ianiro; Sharmila Fagoonee; Carlo R. Settanni; Antonio Gasbarrini; Review: Extragastric Diseases and Helicobacter pylori.. Helicobacter 2020, 25, e12741, doi.org/10.1111/hel.12741.
- Yang Yu; Shengtao Zhu; Peng Li; Li Min; Shutian Zhang; Helicobacter pylori infection and inflammatory bowel disease: a crosstalk between upper and lower digestive tract. Cell Death & Disease 2018, 9, 961, 10.1038/s41419-018-0982-2.
- Shamshul Ansari; Yoshio Yamaoka; Helicobacter pylori Virulence Factors Exploiting Gastric Colonization and its Pathogenicity. Toxins 2019, 11, 677, 10.3390/toxins11110677.
- S. L. Percival; Louise Suleman; Biofilms and Helicobacter pylori: Dissemination and persistence within the environment and host. World Journal of Gastrointestinal Pathophysiology 2014, 5, 122-132, 10.4291/wjgp.v5.i3.122.
- Skander Hathroubi; Julia Zerebinski; Karen M. Ottemann; Regine Hengge; Timothy Cover; Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. mBio 2018, 9, e01973-18, 10.1128/mbio.01973-18.
- Skander Hathroubi; Shuai Hu; Karen M. Ottemann; Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. npj Biofilms and Microbiomes 2020, 6, 1-14, 10.1038/s41522-020-00167-3.
- Chiu-Lien Hung; Hsin-Hung Cheng; Wan-Chen Hsieh; Zing Tsai; Huai-Kuang Tsai; Chia-Han Chu; Wen-Ping Hsieh; Yi-Fan Chen; Yi-Huan Tsou; Chih-Ho Lai; et al.Wen-Ching Wang The CrdRS two-component system in Helicobacter pylori responds to nitrosative stress. Molecular Microbiology 2015, 97, 1128-1141, 10.1111/mmi.13089.
- Miguel A. De La Cruz; Miguel A. Ares; Kristine Von Bargen; Leonardo G. Panunzi; Jessica Martínez-Cruz; Hilda A. Valdez-Salazar; César Jiménez-Galicia; Javier Torres; Gene Expression Profiling of Transcription Factors of Helicobacter pylori under Different Environmental Conditions. Frontiers in Microbiology 2017, 8, 615, 10.3389/fmicb.2017.00615.
- Eric Hong Jian Wong; Chow Goon Ng; Eng-Guan Chua; Alfred Chin Yen Tay; Fanny Peters; Barry J. Marshall; Bow Ho; Khean Lee Goh; Jamuna Vadivelu; Mun Fai Loke; et al. Comparative Genomics Revealed Multiple Helicobacter pylori Genes Associated with Biofilm Formation In Vitro. PLOS ONE 2016, 11, e0166835, 10.1371/journal.pone.0166835.
- Stephanie L. Servetas; Ryan S. Doster; Aeryun Kim; Ian H. Windham; Jeong-Heon Cha; Jennifer A. Gaddy; D Scott Merrell; ArsRS-Dependent Regulation of homB Contributes to Helicobacter pylori Biofilm Formation. Frontiers in Microbiology 2018, 9, 1497, 10.3389/fmicb.2018.01497.
- Hideo Yonezawa; Takako Osaki; Fuhito Hojo; Shigeru Kamiya; Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microbial Pathogenesis 2019, 132, 100-108, 10.1016/j.micpath.2019.04.030.
- Yuying Cai; Caixia Wang; Zhenghong Chen; ZhengZheng Xu; Huanjie Li; Wenjuan Li; Yundong Sun; Transporters HP0939, HP0497, and HP0471 participate in intrinsic multidrug resistance and biofilm formation in Helicobacter pylori by enhancing drug efflux. Helicobacter 2020, 25, e12715, 10.1111/hel.12715.
- Xiaoran Ge; Yuying Cai; Zhenghong Chen; Sizhe Gao; Xiwen Geng; Ya Li; Jihui Jia; Yundong Sun; Yan Li; Bifunctional enzyme SpoT is involved in biofilm formation of Helicobacter pylori with multidrug resistance by upregulating efflux pump Hp1174 (gluP). Antimicrobial Agents and Chemotherapy 2018, 62, 321026, 10.1101/321026.
- Hideo Yonezawa; Takako Osaki; Tomoko Hanawa; Satoshi Kurata; Kuniyasu Ochiai; Shigeru Kamiya; Impact of Helicobacter pylori Biofilm Formation on Clarithromycin Susceptibility and Generation of Resistance Mutations. PLOS ONE 2013, 8, e73301, 10.1371/journal.pone.0073301.
- Bahareh Attaran; T Falsafi; Nassim Ghorbanmehr; Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics.. World Journal of Gastroenterology 2017, 23, 1163-1170, 10.3748/wjg.v23.i7.1163.
- Chunhong Shao; Yundong Sun; Na Wang; Han Yu; Yabin Zhou; Chunyan Chen; Jihui Jia; Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Molecular Medicine Reports 2013, 8, 1761-1766, 10.3892/mmr.2013.1712.
- Feng-Ling Yang; Ammar Mansoor Hassanbhai; Hong-Yu Chen; Zih-You Huang; Tzu-Lung Lin; Shih‐Hsiung Wu; Bow Ho; Proteomannans in Biofilm of Helicobacter pylori ATCC 43504. Helicobacter 2011, 16, 89-98, 10.1111/j.1523-5378.2010.00815.x.
- María G. Cárdenas-Mondragón; Miguel A. Ares; Leonardo G. Panunzi; Sabino Pacheco; Margarita Camorlinga-Ponce; Jorge A. Girón; Javier Torres; Miguel A. De La Cruz; Transcriptional Profiling of Type II Toxin–Antitoxin Genes of Helicobacter pylori under Different Environmental Conditions: Identification of HP0967–HP0968 System. Frontiers in Microbiology 2016, 7, 1872, 10.3389/fmicb.2016.01872.
- Eric Hong Jian Wong; Chow Goon Ng; Khean Lee Goh; Jamuna Vadivelu; Bow Ho; Mun Fai Loke; Metabolomic analysis of low and high biofilm-forming Helicobacter pylori strains. Scientific Reports 2018, 8, 1-9, 10.1038/s41598-018-19697-0.
- Lucinda J. Bessa; Rossella Grande; Donato Di Iorio; Mara Di Giulio; Emanuela Di Campli; Luigina Cellini; Helicobacter pylorifree-living and biofilm modes of growth: behavior in response to different culture media. APMIS 2012, 121, 549-560, 10.1111/apm.12020.
- Emily G. Sweeney; Andrew Nishida; Alexandra Weston; Maria S. Bañuelos; Kristin Potter; John Conery; Karen Guillemin; Agent-Based Modeling Demonstrates How Local Chemotactic Behavior Can Shape Biofilm Architecture. mSphere 2019, 4, e00285-19, 10.1128/msphere.00285-19.
- Luigina Cellini; R. Grande; Tonino Traini; Emanuela Di Campli; S. Di Bartolomeo; D. Di Iorio; S. Caputi; Biofilm formation and modulation of luxS and rpoD expression by Helicobacter pylori. Biofilms 2005, 2, 119-127, 10.1017/s1479050505001845.
- Luigina Cellini; R. Grande; Emanuela Di Campli; S. Di Bartolomeo; Mara Di Giulio; Tonino Traini; O. Trubiani; Characterization of an Helicobacter pylori environmental strain. Journal of Applied Microbiology 2008, 105, 761-769, 10.1111/j.1365-2672.2008.03808.x.
- Indra Bervoets; Daniel Charlier; Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiology Reviews 2019, 43, 304-339, 10.1093/femsre/fuz001.
- Enrique Balleza; Lucia Nikolaia Lopezbojorquez; Agustino Martínez-Antonio; Osbaldo Resendis-Antonio; Irma Lozada-Chávez; Yalbi I. Balderas-Martínez; Sergio Encarnación; Julio Collado-Vides; Regulation by transcription factors in bacteria: beyond description. FEMS Microbiology Reviews 2008, 33, 133-151, 10.1111/j.1574-6976.2008.00145.x.
- Crystal M. Austin; Ge Wang; Robert J. Maier; Aconitase Functions as a Pleiotropic Posttranscriptional Regulator in Helicobacter pylori. Journal of Bacteriology 2015, 197, 3076-3086, 10.1128/jb.00529-15.
- Ge Wang; Judith Romero-Gallo; Stéphane L. Benoit; M. Blanca Piazuelo; Ricardo L. Dominguez; Douglas R. Morgan; Richard M. Peek Jr.; Robert J. Maier; Hydrogen Metabolism in Helicobacter pylori Plays a Role in Gastric Carcinogenesis through Facilitating CagA Translocation. mBio 2016, 7, e01022-16, 10.1128/mbio.01022-16.
- Lisa G. Kuhns; Stéphane L. Benoit; Krishnareddy Bayyareddy; Darryl Johnson; Ron Orlando; Alexandra L. Evans; Grover L. Waldrop; Robert J. Maier; Carbon Fixation Driven by Molecular Hydrogen Results in Chemolithoautotrophically Enhanced Growth of Helicobacter pylori. Journal of Bacteriology 2016, 198, 1423-1428, 10.1128/jb.00041-16.
- Nora-Johanna Krüger; Marie-Theres Knüver; Anna Zawilak-Pawlik; Bernd Appel; Kerstin Stingl; Genetic Diversity as Consequence of a Microaerobic and Neutrophilic Lifestyle. PLOS Pathogens 2016, 12, e1005626, 10.1371/journal.ppat.1005626.
- Davide Roncarati; Alberto Danielli; Gunther Spohn; Isabel Delany; Vincenzo Scarlato; Transcriptional Regulation of Stress Response and Motility Functions in Helicobacter pylori Is Mediated by HspR and HrcA. Journal of Bacteriology 2007, 189, 7234-7243, 10.1128/jb.00626-07.
- Davide Roncarati; Eva M. Pinatel; Elisabetta Fiore; Clelia Peano; Stefany Ojaimi Loibman; Vincenzo Scarlato; Helicobacter pylori Stress-Response: Definition of the HrcA Regulon. Microorganisms 2019, 7, 436, 10.3390/microorganisms7100436.
- Roman Häuser; Markus Pech; Jaroslaw Kijek; Hiroshi Yamamoto; Björn Titz; Florian Naeve; Andrey Tovchigrechko; Kaori Yamamoto; Witold Szaflarski; Nono Takeuchi; et al.Thorsten StellbergerMarkus E. DiefenbacherKnud H. NierhausPeter Uetz RsfA (YbeB) Proteins Are Conserved Ribosomal Silencing Factors. PLOS Genetics 2012, 8, e1002815, 10.1371/journal.pgen.1002815.
- Stephanie L. Servetas; Beth M. Carpenter; Kathryn P. Haley; Jeremy J. Gilbreath; Jennifer A. Gaddy; D Scott Merrell; Characterization of Key Helicobacter pylori Regulators Identifies a Role for ArsRS in Biofilm Formation. Journal of Bacteriology 2016, 198, 2536-2548, 10.1128/jb.00324-16.
- Catherine R. Acio-Pizzarello; Abigail A. Acio; Edward J. Choi; Kimberly Bond; June Kim; Anna Kenan; Jiajia Chen; Mark H. Forsyth; Determinants of the regulation of Helicobacter pylori adhesins include repeat sequences in both promoter and coding regions as well as the two-component system ArsRS. Journal of Medical Microbiology 2017, 66, 798-807, 10.1099/jmm.0.000491.