
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ajaybabu Pobbati | + 1063 word(s) | 1063 | 2020-12-29 04:58:08 | | | |
| 2 | Nicole Yin | Meta information modification | 1063 | 2021-01-11 07:10:02 | | |
Video Upload Options
This a entry that comprehensively covers the modalities that act as disruptors of the YAP-TEAD interaction. The transcriptional co-activator YAP (Yes-associated protein) by pairing with the transcription factor TEAD (TEA domain) orchestrates the expression of several oncogenic transcriptional programs. These programs are seen in a proportion of all solid tumors. Therefore, the disruption of YAP-TEAD interaction is proposed as an attractive option to target cancers.
1. Introduction
The TEADs are a family of four conserved proteins (TEAD 1-4 in humans) that have similar domain architecture (Figure 1A)[1][2]. All TEADs have an N-terminal TEA domain that binds to DNA and adopts a homeodomain fold. At the C-terminus, all TEADs have a transactivation domain that binds to YAP/TAZ. TEADs differ largely at the linker that connects the TEA domain with the transactivation domain. It was known early on that an N-terminal motif of YAP interacts with the TEAD transactivation domain (Figures 1A and 1B). However, the determination of the crystal structures of TEADs in complex with YAP was a breakthrough as it gave a clear, detailed picture of the interaction[3][4][5]. The TEAD transactivation domain adopts an immunoglobulin-like β-sandwich fold; additionally, it has two helix-turn-helix motifs. The N-terminal motif of YAP wraps around TEAD extensively and forms three interfaces (Figure 1B).
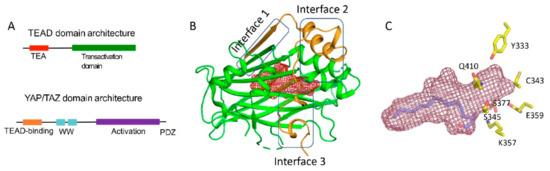
Figure 1. Domain architecture and structures of YAP/TAZ and TEAD (A) Domain architecture of YAP and TEAD (B) Ribbon diagram of the core complex structure of the YAP-TEAD complex (PDB ID: 3KYS). N-terminus of YAP interacts with the transactivation domain of TEAD by forming three interfaces. The central pocket is shown as red mesh. (C) Central pocket (red) and the orientation of the palmitate in the pocket is shown. The conserved polar/charged TEAD2 residues lining the central pocket is shown as yellow sticks.
2. Structural Characterization of YAP-TEAD Interaction
Interface 1 is an anti-parallel β-sheet where both YAP and TEAD contribute to a single β-strand. This interface contributes little to the affinity between YAP and TEAD. Although there are drugs that target higher-order oligomers of β-sheets such as the amyloid plaques formed in Alzheimer disease[6], a simple β-sheet as in interface 1 has poor ligand-binding ability. Interface 2 is formed when a helix of YAP/TAZ slots into a pocket formed by one of the helix-turn-helix motifs of TEAD (Figure 1B). Such a helix-pocket interaction commonly occurs between proteins and is usually mediated by leucine residues (the LxxLL motif)[7][8]. These three hydrophobic leucine residues that are oriented towards the pocket are the key contact residues. YAP and TAZ have a variant of this motif, they have L65xxL68F69 motif instead of LxxLL (the numbers correspond to human YAP residues). Peptides and small molecules have been shown to occupy the interface 2 pocket (Table 1). The YAP/TAZ helix on its own binds poorly to TEAD and interface 3 acts to increase its binding affinity. Interface 3 is the most crucial of the three interfaces[3][4][5]. At this interface, YAP adopts a three-dimensional structure resembling the Greek letter Ω (Figure 1B), and this conformation is generally referred to as the Ω-loop. Crucial for interface 3 interaction are three hydrophobic core residues -- in human YAP, these are M86, L91, and F95 -- that fit into a pocket on the TEAD surface. A cation-π interaction formed between R87 and F96 YAP residues is also crucial and appears to stabilize the Ω-loop conformation. Appreciable progress has been made on the peptidomimetics front, linear and cyclic peptides have been designed to act as PPIDs by occupying the interface 3 pocket on TEAD (Table 1). Interface 3 TEAD residues that interact with YAP are highly conserved, therefore it is difficult to design modalities at this interface that display selective TEAD inhibition. That said, it is not clear whether there is a biological need for selective TEAD inhibitors. In the field, researchers tend to work more on YAP than TAZ, but as YAP and TAZ have similar primary sequences and structural features, PPIDs that disrupt the YAP-TEAD interaction should also have the ability to disrupt the TAZ-TEAD interaction.
Table 1. Compounds that target TEAD surface pocket interfaces 2 and 3.
| No. | Molecule | Structure | Surface Pocket Validation | Molecule Type | Validation Method | Reference |
|---|---|---|---|---|---|---|
| Modalities binding to Interface 2 | ||||||
| 1. | TB2G1 | Cystine-dense peptide | Rosetta modeling | PPID | Co-IP assay | [9] |
| 2. | Fragment 1 | 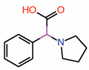 |
Crystal structure | PPID unlikely |
GST pull-down | [10] |
| 3. | Tri-substitutedpyrazoles | 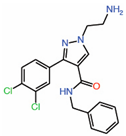 |
Crystal structure | Potential PPID |
||
| Modalities binding to Interface 3 | ||||||
| 4. | Peptide 17 | YAP cyclic peptide | Molecular modeling | PPID | GST pull-down | [11] |
| 5. | Peptide 10 | YAP cyclic peptide | Crystal structure |
PPID | GST pull-down | [12] |
| 6. | Peptides 9, 10 | YAP linear peptide | Crystal structure |
PPID | TR-FRET assay | [13] |
| 7. | Flufenamic acid |  |
Crystal structure |
PPID unlikely |
[14] | |
| 8. | TEAD-binding fragment |
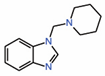 |
NMR | PPID unlikely |
[15] | |
| 9. | Dioxo-benzoisothiazole Example 22 |
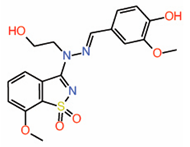 |
NMR | PPID | AlphaLISA assay |
[16] |
| 10. | Triazole carbohydrazides Hit 2 |
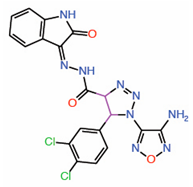 |
Molecular modeling |
Potential PPID |
[17] | |
| 11. | Compound 3.1 | 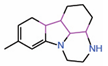 |
Molecular docking |
PPID | Co-IP assay | [18] |
| Peptide binding to both Interfaces 2 & 3 | ||||||
| 12. | Super-TDU | YAP-VGLL4 fusion peptide | Molecular modeling |
PPID | Co-IP assay | [19] |
Interestingly, Pobbati and colleagues identified a pocket with an appropriate geometry and hydrophobicity in the center of the transactivating domain of TEADs that is accessible to small molecules[14] (Figure 1B). This pocket is distinct from the TEAD surface pockets. One end of the pocket is solvent-exposed. However, it is blocked by the interface 1 β-strand of YAP that acts as a gate to ligand access. The other end of the central pocket extends into the domain interior. Palmitate is the natural ligand that binds to the central pocket[20][21]. In addition to the hydrophobic residues that line the TEAD central pocket, there are seven conserved charged/polar residues (Figure 1C). Remarkably, all seven residues are located at the solvent-exposed end of the pocket. Notably, several central pocket binders have been identified that, not only inhibit TEAD palmitoylation, but also disrupt YAP-TEAD interaction, thus, acting as allosteric PPIDs. Several other molecules bind the pocket but only act as palmitoylation inhibitors and not as allosteric PPIDs. Despite this, they inhibit TEAD activity and function (Table 2).
Table 2. Compounds that target the TEAD central pocket.
| No. | Molecule | Structure | Binding Validation | Molecule Type | Validation Method | Reference |
|---|---|---|---|---|---|---|
| Flufenamates | ||||||
| 1. | Flufenamic acid | 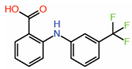 |
Crystal structure | Palmitoylation inhibitor |
Mass Spec analysis | [14] |
| 2. | TED-347 | 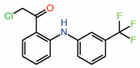 |
Crystal structure | Allosteric PPID |
FP assay with YAP peptide | [22] |
| 3. | MYF-01-037 | 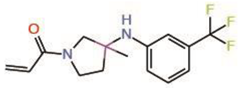 |
Molecular docking |
Allosteric PPID |
Split luciferase assay | [23] |
| 4. | Non-fused tricyclic Compound 42 | 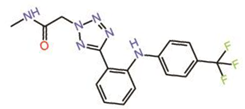 |
unknown | PPID? | [24] | |
| 5. | FA-Palmitate fusion | 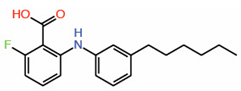 |
Crystal structure | PPID? | ||
| Non-covalent allosteric PPIDs | ||||||
| 6. | MGH-CP1 | 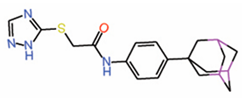 |
Crystal structure | Allosteric PPID |
Co-IP assay | [25] |
| 7. | Compound 9 Indole-focused | 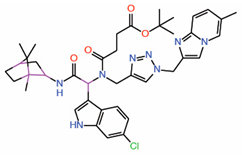 |
Palmitate-based FP assay | Allosteric PPID |
FP assay with YAP peptide | [26] |
| 8. | Dihydropyrazolo pyrimidines Compound 7 |
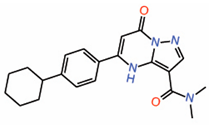 |
unknown | AllostericPPID? | TR-FRET assay | [27] |
| Covalent allosteric PPIDs | ||||||
| 9. | K-975 | 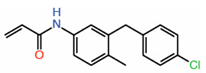 |
Crystal structure | Allosteric PPID |
Co-IP assay | [28] |
| 10. | Kojic acid analogs Compound 19 | 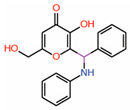 |
Thiol conjugation assay | Allosteric PPID |
FP assay with YAP peptide | [29] |
| Other central pocket binders | ||||||
| 11. | Compound 2 | 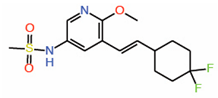 |
Crystal structure | Palmitoylation inhibitor | FP assay with palmitate | [30] |
| 12. | Vinylsulfonamide DC-TEADin02 | 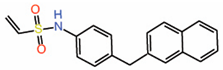 |
Molecular docking | Palmitoylation inhibitor | Competitive NMR |
[31] |
| 13. | Quinolinol Q2 |  |
Molecular dynamics simulations |
TEAD activator | RNA-Seq | [32] |
References
- Pobbati, A.V.; Hong, W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol. 2013, 14, 390–398.
- Huh, H.D.; Kim, D.H.; Jeong, H.S.; Park, H.W. Regulation of TEAD Transcription Factors in Cancer Biology. Cells 2019, 8, 600.
- Wei Tian; Jianzhong Yu; Diana R. Tomchick; Duojia Pan; Xuelian Luo; Structural and functional analysis of the YAP-binding domain of human TEAD2. Proceedings of the National Academy of Sciences 2010, 107, 7293-7298, 10.1073/pnas.1000293107.
- Li, Z.; Zhao, B.; Wang, P.; Chen, F.; Dong, Z.; Yang, H.; Guan, K.L.; Xu, Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010, 24, 235–240.
- Chen, L.; Chan, S.W.; Zhang, X.; Walsh, M.; Lim, C.J.; Hong, W.; Song, H. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010, 24, 290–300.
- Li-Kai Huang; Shu-Ping Chao; Chaur-Jong Hu; Clinical trials of new drugs for Alzheimer disease. Journal of Biomedical Science 2020, 27, 1-13, 10.1186/s12929-019-0609-7.
- Darimont, B.D.; Wagner, R.L.; Apriletti, J.W.; Stallcup, M.R.; Kushner, P.J.; Baxter, J.D.; Fletterick, R.J.; Yamamoto, K.R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998, 12, 3343–3356.
- Nolte, R.T.; Wisely, G.B.; Westin, S.; Cobb, J.E.; Lambert, M.H.; Kurokawa, R.; Rosenfeld, M.G.; Willson, T.M.; Glass, C.K.; Milburn, M.V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 1998, 395, 137–143.
- Zachary R. Crook; Gregory P. Sevilla; Della Friend; Mi-Youn Brusniak; Ashok D. Bandaranayake; Midori Clarke; Mesfin Gewe; Andrew J. Mhyre; David Baker; Roland K. Strong; et al.Philip BradleyJames M. Olson Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets. Nature Communications 2017, 8, 2244, 10.1038/s41467-017-02098-8.
- Hung Yi Kristal Kaan; Adelene Y. L. Sim; Siew Kim Joyce Tan; Chandra S. Verma; Haiwei Song; Targeting YAP/TAZ-TEAD protein-protein interactions using fragment-based and computational modeling approaches. PLOS ONE 2017, 12, e0178381, 10.1371/journal.pone.0178381.
- Zhisen Zhang; Zhaohu Lin; Zheng Zhou; Hong C. Shen; S. Frank Yan; Alexander V. Mayweg; Zhiheng Xu; Ning Qin; Jason C. Wong; Zhenshan Zhang; et al.Yiping RongDavid C. FryTaishan Hu Structure-Based Design and Synthesis of Potent Cyclic Peptides Inhibiting the YAP–TEAD Protein–Protein Interaction. ACS Medicinal Chemistry Letters 2014, 5, 993-998, 10.1021/ml500160m.
- Zheng Zhou; Taishan Hu; Zhiheng Xu; Zhaohu Lin; Zhisen Zhang; Teng Feng; Liangcheng Zhu; Yiping Rong; Hong Shen; John M. Luk; et al.Xiongwen ZhangNing Qin Targeting Hippo pathway by specific interruption of YAP‐TEAD interaction using cyclic YAP‐like peptides. The FASEB Journal 2014, 29, 724-732, 10.1096/fj.14-262980.
- Pascal Furet; Bahaa Salem; Yannick Mesrouze; Tobias Schmelzle; Ian Lewis; Joerg Kallen; Patrick Chène; Structure-based design of potent linear peptide inhibitors of the YAP-TEAD protein-protein interaction derived from the YAP omega-loop sequence. Bioorganic & Medicinal Chemistry Letters 2019, 29, 2316-2319, 10.1016/j.bmcl.2019.06.022.
- Ajaybabu V. Pobbati; Xiao Han; Alvin W. Hung; Seetoh Weiguang; Nur Huda; Guo-Ying Chen; Congbao Kang; Cheng San Brian Chia; Xuelian Luo; Wanjin Hong; et al.Anders Poulsen Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure 2015, 23, 2076-2086, 10.1016/j.str.2015.09.009.
- Yan Li; Shuang Liu; Elizabeth Yihui Ng; Rong Li; Anders Poulsen; Jeffrey Hill; Ajaybabu V. Pobbati; Alvin W. Hung; Wanjin Hong; Thomas H. Keller; et al.Congbao Kang Structural and ligand-binding analysis of the YAP-binding domain of transcription factor TEAD4. Biochemical Journal 2018, 475, 2043-2055, 10.1042/bcj20180225.
- Barth, M.C.S.; Montalbetti, C.; Spitzer, L. Preparation of new 4-(1,1-Dioxo-1,2-benzothiazol-3-yl)hydrazono]methyl]-2-methoxyphenols as Inhibitors of the YAP/TAZ-TEAD Interaction and Their Use in the Treatment of Malignant Mesothelioma. WO Patent 2017064277, 20 April 2017. Inventiva.
- Floriane Gibault; Mathilde Coevoet; Manon Sturbaut; Amaury Farce; Pascal Carato; Frédéric Allemand; Jean-François Guichou; Anne-Sophie Drucbert; Catherine Foulon; Romain Magnez; et al.Xavier ThuruMatthieu CorvaisierGuillemette HuetPhilippe ChavattePatricia MelnykFabrice BaillyPhilippe Cotelle Toward the Discovery of a Novel Class of YAP–TEAD Interaction Inhibitors by Virtual Screening Approach Targeting YAP–TEAD Protein–Protein Interface. Cancers 2018, 10, 140, 10.3390/cancers10050140.
- Sarah A. Smith; Richard B. Sessions; Deborah K. Shoemark; Christopher Williams; Reza Ebrahimighaei; Madeleine C. McNeill; Matthew P. Crump; Tristan R. McKay; Gemma Harris; A. C. Newby; et al.Mark Bond Antiproliferative and Antimigratory Effects of a Novel YAP–TEAD Interaction Inhibitor Identified Using in Silico Molecular Docking. Journal of Medicinal Chemistry 2019, 62, 1291-1305, 10.1021/acs.jmedchem.8b01402.
- Shi Jiao; Huizhen Wang; Zhubing Shi; Aimei Dong; Wenjing Zhang; Xiaomin Song; Feng He; Yicui Wang; Zhenzhen Zhang; Wenjia Wang; et al.Xin WangTong GuoPeixue LiYun ZhaoHongBin JiLei ZhangZhaocai Zhou A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer. Cancer Cell 2014, 25, 166-180, 10.1016/j.ccr.2014.01.010.
- Chan, P.; Han, X.; Zheng, B.; DeRan, M.; Yu, J.; Jarugumilli, G.K.; Deng, H.; Pan, D.; Luo, X.; Wu, X. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat. Chem. Biol. 2016, 12, 282–289.
- Noland, C.L.; Gierke, S.; Schnier, P.D.; Murray, J.; Sandoval, W.N.; Sagolla, M.; Dey, A.; Hannoush, R.N.; Fairbrother, W.J.; Cunningham, C.N. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure 2016, 24, 179–186.
- Khuchtumur Bum-Erdene; Donghui Zhou; Giovanni Gonzalez-Gutierrez; Mona K. Ghozayel; Yubing Si; David Xu; Harlan E. Shannon; Barbara J. Bailey; Timothy W. Corson; Karen E. Pollok; et al.Clark D. WellsSamy O. Meroueh Small-Molecule Covalent Modification of Conserved Cysteine Leads to Allosteric Inhibition of the TEAD⋅Yap Protein-Protein Interaction. Cell Chemical Biology 2019, 26, 378-389.e13, 10.1016/j.chembiol.2018.11.010.
- Jean Clairambault; Kurppa Kj; Liu Y; To C; Zhang T; Fan M; Vajdi A; Knelson Eh; Xie Y; Lim K; et al.Cejas PPortell ALizotte PhFicarro SbLi SChen THaikala HmWang HBahcall MGao YShalhout SBoettcher SShin BhThai TWilkens MkTillgren MlMushajiang MXu MChoi JBertram AaEbert BlBeroukhim RBandopadhayay PAwad MmGokhale PcKirschmeier PtMarto JaCamargo FdHaq RPaweletz CpWong KkBarbie DaLong HwGray NsJänne Pa Faculty Opinions recommendation of Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway.. Faculty Opinions – Post-Publication Peer Review of the Biomedical Literature 2020, 37, 104–122 e12., 10.3410/f.737201348.793575388.
- Konradi, A.W.; Lin, T.T.L.T. Non-Fused Tricyclic Compounds. WO Patent 2018204532, 8 November 2018. Vivace Therapeutics.
- Qi Li; Yang Sun; Gopala K. Jarugumilli; Shun Liu; Kyvan Dang; Jennifer L. Cotton; Jordi Xiol; Pui Yee Chan; Michael DeRan; Lifang Ma; et al.Rui LiLihua J. ZhuJoyce H. LiAndrew B. LeiterY. Tony IpFernando D. CamargoXuelian LuoRandy L. JohnsonXu WuJunhao Mao Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription. Cell Stem Cell 2020, 26, 675-692.e8, 10.1016/j.stem.2020.03.002.
- Verena B. K. Kunig; Marco Potowski; Mohammad Akbarzadeh; Mateja Klika Škopić; Denise Dos Santos Smith; Lukas Arendt; Ina Dormuth; Hélène Adihou; Blaž Andlovic; Hacer Karatas; et al.Shabnam ShaabaniTryfon Zarganes‐TzitzikasDr. Constantinos G. NeochoritisRan ZhangDr. Matthew GrovesStéphanie M. GuéretChristian OttmannDr. Jörg RahnenführerDr. Roland FriedDr. Alexander DömlingAndreas Brunschweiger TEAD–YAP Interaction Inhibitors and MDM2 Binders from DNA‐Encoded Indole‐Focused Ugi Peptidomimetics. Angewandte Chemie 2020, 132, 20518-20522, 10.1002/ange.202006280.
- Zbieg, P.P.; Crawford, J.J. Therapeutic Compounds. WO Patent 2019232216, 22 August 2019.
- Ayumi Kaneda; Toshihiro Seike; Takeshi Uemori; Kensuke Myojo; Kensuke Aida; Tomohiro Danjo; Takahiro Nakajima; Daisuke Yamaguchi; Tomoko Hamada; Yoshiro Tsuji; et al.Kaori HamaguchiMai YasunagaNobumasa OtsuboHideyuki OnoderaYoichi NishiyaMichihiko SuzukiJunichi SaitoToshihiko IshiiRyuichiro Nakai Abstract 3086: Discovery of afirst-in-classTEAD inhibitor which directly inhibits YAP/TAZ-TEAD protein-protein interaction and shows a potent anti-tumor effect in malignant pleural mesothelioma. Experimental and Molecular Therapeutics 2019, 79, 3086-3086, 10.1158/1538-7445.sabcs18-3086.
- Hacer Karatas; Mohammad Akbarzadeh; Hélène Adihou; Gernot Hahne; Ajaybabu V. Pobbati; Elizabeth Yihui Ng; Stéphanie M. Guéret; Sonja Sievers; Axel Pahl; Malte Metz; et al.Sarah ZinkenLara DötschChristine NowakSasikala ThavamAlexandra FrieseCongbao KangWanjin HongDr. Herbert Waldmann Discovery of Covalent Inhibitors Targeting the Transcriptional Enhanced Associate Domain Central Pocket. Journal of Medicinal Chemistry 2020, 63, 11972-11989, 10.1021/acs.jmedchem.0c01275.
- Jeffrey K. Holden; James J. Crawford; Cameron L. Noland; Stephen Schmidt; Jason R. Zbieg; Jennifer A. Lacap; Richard Zang; Gregory M. Miller; Yue Zhang; Paul Beroza; et al.Rohit RejaWendy LeeJeffrey Y.K. TomRina FongMicah SteffekSaundra ClausenThjis J. HagenbeekTaishan HuZheng ZhouHong C. ShenChristian N. Cunningham Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling. Cell Reports 2020, 31, 107809, 10.1016/j.celrep.2020.107809.
- Wenchao Lu; Jun Wang; Yong Li; Hongru Tao; Huan Xiong; Fulin Lian; Jing Gao; Hongna Ma; Tian Lu; Dan Zhang; et al.Xiaoqing YeHong DingLiyan YueYuanyuan ZhangHuanyu TangNaixia ZhangYaxi YangHualiang JiangKaixian ChenBing ZhouCheng Luo Discovery and biological evaluation of vinylsulfonamide derivatives as highly potent, covalent TEAD autopalmitoylation inhibitors. European Journal of Medicinal Chemistry 2019, 184, 111767, 10.1016/j.ejmech.2019.111767.
- Ajaybabu V. Pobbati; Tom Mejuch; Sayan Chakraborty; Hacer Karatas; Sakshibeedu R. Bharath; Stéphanie M. Guéret; Pierre-Alexis Goy; Gernot Hahne; Axel Pahl; Sonja Sievers; et al.Ernesto GuccioneHaiwei SongDr. Herbert WaldmannWanjin Hong Identification of Quinolinols as Activators of TEAD-Dependent Transcription. ACS Chemical Biology 2019, 14, 2909-2921, 10.1021/acschembio.9b00786.




