| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Katherine L. B. Borden | + 2868 word(s) | 2868 | 2020-12-29 09:58:30 | | | |
| 2 | Rita Xu | -1008 word(s) | 1860 | 2021-01-08 03:24:58 | | |
Video Upload Options
Export of mRNAs from the nucleus to the cytoplasm is a key regulatory step in the expression of proteins. mRNAs are transported through the nuclear pore complex (NPC) in association with key co-factors. Export of selected subsets of mRNAs permits targeted elevation of specific protein expression. Indeed, there are different export pathways depending on the mRNA underpinning this selectivity. Export of mRNAs can respond to a variety of cellular stimuli making it a dynamic means to respond to extracellular stimuli. mRNA export can be dysregulated in, and contribute to, human diseases including cancer and neurodegeneration. This entry focuses on its role in cancer.
1. Overview
Dysregulation of many cellular processes contributes to cancer. Aberrant transcription, translation, and signaling are amongst the most widely studied. However, the entire journey of coding RNA from transcription in the nucleus to translation into protein in the cytoplasm is marked by important RNA processing steps such as m7G capping, splicing, polyadenylation, and subsequent export to the cytoplasm [1][2][3][4][5][6][7]. Indeed, the transcriptome does not always predict the proteome [8] this arises in large part because of regulation at the post-transcriptional level, which can decouple transcription and translation. Not surprisingly, many aspects of RNA processing including RNA export are now known to be dysregulated in, and contribute to, cancer [1][2][3][4][5][6][7]. To be exported, RNAs associate with a wide array of adaptor proteins and are exported as large ribonucleotide complexes (mRNPs), which enable them to traverse the channel in the nuclear envelope known as the nuclear pore complex (NPC) [4][6][7]. Indeed, RNA export factors and NPC components regulate the export of selected RNAs that act in nearly all facets of malignancy, e.g., survival, proliferation, metastases, and invasion [4][6][7]. Many signaling pathways converge on the RNA export machinery as well as the NPC, thereby positioning this process as a mediator of signaling and stress responses [4][6][7].
2. General Features of Nuclear-Cytoplasmic Trafficking
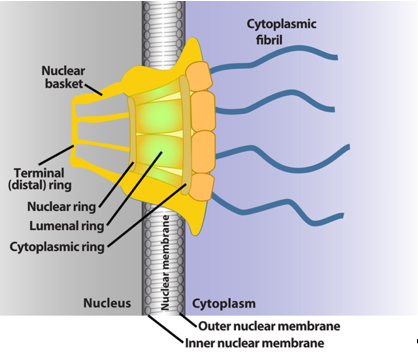
The NPC is a megadalton protein complex embedded within the nuclear membrane and serves as the primary transit route between the nucleus and cytoplasm. This route is used by a wide array of macromolecules including mRNA. The structure of the NPC was first described in early EM studies as an apparently hollow cylindrical moiety embedded in the nuclear membrane with an eightfold symmetry around the pore [9][10]. The NPC has been the subject of intense structural studies and its basic structural features are conserved from fungi to humans [11][12][13][14] (and references therein). High-resolution cryo-electron tomography has been used to generate 3D reconstructions of intact NPCs while NMR and X-ray crystallography have been employed to elucidate the structures of subcomplexes at atomic resolution [11][12][13]. These studies reveal that the NPC is composed of three main structural features: a nuclear basket, a central membrane-traversing channel, and cytoplasmic fibrils (Figure 1). The nuclear basket and cytoplasmic fibrils are attached to the central framework of the pore through nuclear rings and cytoplasmic rings, respectively. The nuclear basket is formed by eight filaments joined by a distal ring, while eight cytoplasmic filaments or fibrils are anchored by the cytoplasmic rings (for clarity of presentation, only four are shown in Figure 1). In humans, the NPC has a molecular mass of ~10 MDa and is comprised of ~30 nucleoporins (Nups), which are present in multiple copies. The protein constituent Nups are attributed names on the basis of their molecular mass. The NPC is characterized with outer and inner diameters of approximately 120 nm and 40 nm, respectively [15][16]. The nuclear basket serves as a docking site for export cargoes with the NPC. The central channel is set within the nuclear envelope and is not hollow. Within the channel, intrinsically disordered phenylalanine–glycine (FG) repeat proteins constitute a diffusion barrier. Macromolecule cargoes smaller than ~30–50 kDa can traverse passively through the diffusion barrier whereas larger molecules require nuclear transport factors (also known as karyopherins) for successful translocation [11][17][18]. Characteristics of cargoes such as their surface charge can dramatically alter their permeability, allowing larger factors to diffuse passively in some instances [19][20][21]. On the cytoplasmic side, filaments known as cytoplasmic fibrils can project up to 50 nm outward into the cytoplasm [11][12][13]. These function as cargo release sites for export cargoes or docking sites for nuclear import [4][22][23][24]. It is important to note that NPC composition can differ in specific cell types, revealing context-dependent transport functions [25][26][27]. Further, the NPC and some associated factors can also act in non-transport functions [25][26][27][28]; however, we will focus on transport activities here.
Most cargoes must associate with nuclear transport receptors and associated factors to transit through the NPC. Cargoes typically interact with nuclear transport receptors such as importins, exportins, or transportins. For import of cargoes into the nucleus, transport typically requires protein cargoes to contain an accessible nuclear localization signals (NLS) [29]. In the cytoplasm, importins associate with cargoes displaying accessible NLSs and allow these to transit through the NPC to the nucleus. Once in the nucleus, cargoes must be released from the importin. This occurs via association of RanGTP with the importin, which induces conformational changes that lead to cargo release. Subsequently the RanGTP–importin complex is recycled to the cytoplasm [29][30][31]. In the cytoplasm, the RanGTPase-activating protein (RanGAP) in the presence of Ran-binding proteins (RanBPs) substantially stimulates hydrolysis of RanGTP, and the subsequent RanGDP dissociates from the cargo for release into the cytoplasm, freeing the importin for future rounds of nuclear import [29].
Figure 1. Schematics of the basic elements of the vertebrate nuclear pore complex. The nuclear basket, central pore, and cytoplasmic fibrils are shown. For simplicity, only four cytoplasmic fibrils and four nuclear basket protrusions are shown, rather than all eight.
Export from the nucleus generally necessitates cargoes to possess an accessible nuclear export signal (NES), permitting association with exportin proteins. The prevalent exportin is known as exportin 1 or chromosome maintenance protein 1 (XPO1/CRM1). Here, cargoes with an accessible NES form a complex with an exportin and RanGTP [32][33][34]. This complex can now associate with factors at the nuclear basket and transit through the central channel of the nuclear pore. At the cytoplasmic side, one of the major components of the fibrils is Nup358, also known as RanBP2 [11][12][13]. RanBP2 contains docking sites for many proteins and binds RanGAP as well as binding sites for both RanGDP and RanGTP [12][13][24][35]. Here, the cargoes are released from the exportin by hydrolysis of RanGTP to RanGDP through RanGAP associated with RanBP2 or in some cases with the small, soluble RanBP1 [36]. Hydrolysis to RanGDP reduces the affinity of the exportin for the NES-containing cargo, allowing its release into the cytoplasm and subsequent recycling of the exportin to the nucleus for future rounds of export [37]. To ensure directionality of transport, Ran is recycled by the nuclear transport factor 2 (NTF2), which binds RanGDP in the cytoplasm and ferries it to the nucleoplasm [36]. Here, the RanGEF exchange factor RCC1 facilitates nucleotide exchange to regenerate RanGTP [36].
3. General Features of mRNA Export
mRNA export is extremely important for gene regulation, serving two critical functions: (1) it provides a surveillance mechanism, meaning that it weeds out aberrant mRNAs, and thus these do not become translated into aberrant proteins with altered functions. (2) It serves as an important regulatory step, permitting altered flow of specific mRNAs into the cytoplasm in order to control their translation into protein and thus the response to extracellular signals as well as stress conditions. Indeed, groups of mRNAs encoding proteins acting in the same biochemical pathways can be coordinately exported, or retained in the nucleus, providing a powerful means to quickly turn on or off biochemical pathways that drive cell physiology [38][39][40][41]. This model of how to control groups of mRNAs is referred to as RNA regulons. This coordinated control typically arises from the presence of cis-acting elements conserved amongst mRNA targets, which are referred to as USER (untranslated sequence elements for regulation) codes [38][39][40][41]. Transit through the NPC is by far the major exit route for export, but there are examples of large mRNPs exiting the nucleus by budding at the nuclear membrane [42]. In this entry, the focus is on the NPC route. As will be described below, the NPC plays an active role in RNA export and indeed its modulation can impact on bulk mRNA export or on selected mRNAs.
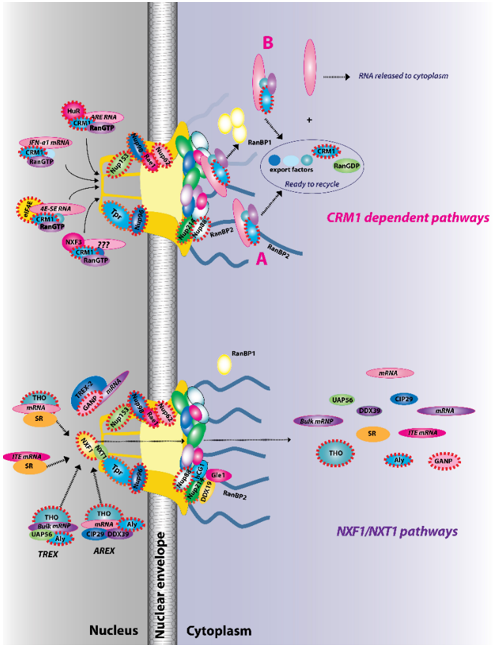
Given the fundamental roles mRNA export plays in gene expression, it is not surprising that RNA export is typically closely tied to RNA processing. For mRNAs, this generally involves the addition of a 7-methylguanosine (m7G cap) on the 5′ end of transcripts, splicing and addition of a polyA tail on the 3′ end of transcripts. Interactions with particular protein factors mark the mRNA as processed and ready for export [4][6]. Generally speaking, mRNA cargoes cannot directly interact with proteins at the nuclear basket. Rather, their protein co-factors influence the ability of cargo mRNPs to form complexes with nuclear receptors, which in turn mediate their interaction with the nuclear basket and subsequent traversal of the NPC [4][6]. For the export of the majority of RNAs, the m7G cap plays a key role permitting interactions with the cap-binding protein complex (CBC) or in some cases with the eukaryotic translation initiation factor eIF4E as well as other factors [6]. To date, the best studied mRNA export receptors are the nuclear RNA export factor 1 (NXF1)/Tip-associating protein (TAP) acting in complex with NXT1 (nuclear transport factor 2-like export 1)/p15 [43][44] or XPO1/CRM1 [4][6] (Figure 2). The majority of mRNAs use the NXF1/NXT1 heterodimer route [4][45]. RNA export can be modulated at several stages, including assembly of mRNPs in the nucleus, association with the nuclear basket, RNA cargo release in the cytoplasm, and recycling of export factors back to the nucleus. This provides multiple steps that can respond to extracellular stimuli, stress, intracellular signals, or can be dysregulated in cancer.
Figure 2. The two major RNA export receptor pathways, nuclear RNA export factor 1 (NXF1)/nuclear transport factor 2-like export 1 (NXT1) and chromosome maintenance protein 1 (CRM1), are shown, and both are dysregulated in cancer. There are multiple routes to engage either of these pathways. For example, for the NXF1/NXT1 pathways, there are complexes that rely on ALY/REF (transcription export (TREX) and AREX) and also on GANP (TREX2). There are the intronless H2a RNAs with intronless transport elements (ITEs) that rely on SR proteins but additionally some intron-containing RNAs also require SR proteins. For the NXF1/NXT1 pathway, cargo release depends on DDX9/Gle1. There are also multiple routes to engage the CRM1 pathways. These are depicted for IFN1α, HuR, eIF4E, and NXF3. For the CRM1 pathway, cargo release involves RanGTP hydrolysis through either the RanBP1 or RanBP2 pathways (A). For the case of eIF4E overexpression (B), RanBP2 levels are reduced (depicted by fibrils in shadow) and RanBP1 levels are increased. Thus, the RanBP1 release pathways are thought to predominate. Once cargoes are released from both the NXF1/NXT1 and CRM1 pathways, exportins and associated factors are recycled (not shown). Only four cytoplasmic fibrils and four nuclear basket protrusions are displayed for simplicity of presentation (as there are eight fibrils per nuclear pore complex (NPC)). Factors known to be involved in cancer are encapsulated in red dashed lines. This is not an exhaustive list of all NPC factors that are dysregulated in cancer but highlights the fact that all facets of the NPC can be impacted (nuclear basket, central channel, and cytoplasmic fibrils).
References
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Rev. Mol. Cell Biol. 2017, 18, 18–30, doi:10.1038/nrm.2016.116.
- Baralle, D.; Buratti, E. RNA splicing in human disease and in the clinic. Sci. (Lond.) 2017, 131, 355–368, doi:10.1042/CS20160211.
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Rev. Mol. Cell Biol. 2017, 18, 437–451, doi:10.1038/nrm.2017.27.
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373, doi:10.1016/j.tcb.2013.03.006.
- Cowling, V.H.; Cole, M.D. Myc Regulation of mRNA Cap Methylation. Genes Cancer 2010, 1, 576–579, doi:10.1177/1947601910378025.
- Culjkovic-Kraljacic, B.; Borden, K.L. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013, 23, 328–335, doi:10.1016/j.tcb.2013.03.004.
- Bonnet, A.; Palancade, B. Regulation of mRNA trafficking by nuclear pore complexes. Genes 2014, 5, 767–791, doi:10.3390/genes5030767.
- de Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Biosyst. 2009, 5, 1512–1526, doi:10.1039/b908315d.
- Watson, M.L. The nuclear envelope; its structure and relation to cytoplasmic membranes. Biophys. Biochem. Cytol. 1955, 1, 257–270, doi:10.1083/jcb.1.3.257.
- Gall, J.G. Octagonal nuclear pores. Cell Biol. 1967, 32, 391–399, doi:10.1083/jcb.32.2.391.
- Field, M.C.; Rout, M.P. Pore timing: The evolutionary origins of the nucleus and nuclear pore complex. F1000Research 2019, 8, doi:10.12688/f1000research.16402.1.
- Lin, D.H.; Hoelz, A. The Structure of the Nuclear Pore Complex (An Update). Rev. Biochem. 2019, 88, 725–783, doi:10.1146/annurev-biochem-062917-011901.
- Hampoelz, B.; Andres-Pons, A.; Kastritis, P.; Beck, M. Structure and Assembly of the Nuclear Pore Complex. Rev. Biophys. 2019, 48, 515–536, doi:10.1146/annurev-biophys-052118-115308.
- Schwartz, T.U. The Structure Inventory of the Nuclear Pore Complex. Mol. Biol. 2016, 428, 1986–2000, doi:10.1016/j.jmb.2016.03.015.
- Kosinski, J.; Mosalaganti, S.; von Appen, A.; Teimer, R.; DiGuilio, A.L.; Wan, W.; Bui, K.H.; Hagen, W.J.; Briggs, J.A.; Glavy, J.S.; et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 2016, 352, 363–365, doi:10.1126/science.aaf0643.
- Lin, D.H.; Stuwe, T.; Schilbach, S.; Rundlet, E.J.; Perriches, T.; Mobbs, G.; Fan, Y.; Thierbach, K.; Huber, F.M.; Collins, L.N.; et al. Architecture of the symmetric core of the nuclear pore. Science 2016, 352, aaf1015, doi:10.1126/science.aaf1015.
- Bonner, W.M. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. Cell Biol. 1975, 64, 421–430, doi:10.1083/jcb.64.2.421.
- Paine, P.L.; Moore, L.C.; Horowitz, S.B. Nuclear envelope permeability. Nature 1975, 254, 109–114, doi:10.1038/254109a0.
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. Cell Biol 2016, 215, 57–76, doi:10.1083/jcb.201601004.
- Naim, B.; Zbaida, D.; Dagan, S.; Kapon, R.; Reich, Z. Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 2009, 28, 2697–2705, doi:10.1038/emboj.2009.225.
- Frey, S.; Rees, R.; Schunemann, J.; Ng, S.C.; Funfgeld, K.; Huyton, T.; Gorlich, D. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell 2018, 174, 202–217 e209, doi:10.1016/j.cell.2018.05.045.
- Bernad, R.; van der Velde, H.; Fornerod, M.; Pickersgill, H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Cell Biol. 2004, 24, 2373–2384, doi:10.1128/mcb.24.6.2373-2384.2004.
- Hamada, M.; Haeger, A.; Jeganathan, K.B.; van Ree, J.H.; Malureanu, L.; Walde, S.; Joseph, J.; Kehlenbach, R.H.; van Deursen, J.M. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. Cell Biol. 2011, 194, 597–612, doi:10.1083/jcb.201102018.
- Hutten, S.; Walde, S.; Spillner, C.; Hauber, J.; Kehlenbach, R.H. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. Cell Sci. 2009, 122, 1100–1110, doi:10.1242/jcs.040154.
- Hezwani, M.; Fahrenkrog, B. The functional versatility of the nuclear pore complex proteins. Cell Dev. Biol. 2017, 68, 2–9, doi:10.1016/j.semcdb.2017.05.004.
- Juhlen, R.; Fahrenkrog, B. Moonlighting nuclear pore proteins: Tissue-Specific nucleoporin function in health and disease. Cell Biol. 2018, 150, 593–605, doi:10.1007/s00418-018-1748-8.
- Sakuma, S.; D’Angelo, M.A. The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Cell Dev. Biol. 2017, 68, 72–84, doi:10.1016/j.semcdb.2017.05.006.
- Chatel, G.; Fahrenkrog, B. Nucleoporins: Leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011, 23, 1555–1562, doi:10.1016/j.cellsig.2011.05.023.
- Chook, Y.M.; Suel, K.E. Nuclear import by karyopherin-betas: Recognition and inhibition. Biophys. Acta 2011, 1813, 1593–1606, doi:10.1016/j.bbamcr.2010.10.014.
- Chook, Y.M.; Blobel, G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 1999, 399, 230–237, doi:10.1038/20375.
- Gorlich, D. Transport into and out of the cell nucleus. EMBO J. 1998, 17, 2721–2727, doi:10.1093/emboj/17.10.2721.
- Dong, X.; Biswas, A.; Suel, K.E.; Jackson, L.K.; Martinez, R.; Gu, H.; Chook, Y.M. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 2009, 458, 1136–1141, doi:nature07975 [pii]10.1038/nature07975.
- Monecke, T.; Guttler, T.; Neumann, P.; Dickmanns, A.; Gorlich, D.; Ficner, R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 2009, 324, 1087–1091, doi:10.1126/science.1173388.
- Dong, X.; Biswas, A.; Chook, Y.M. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Struct Mol. Biol 2009, 16, 558–560, doi:10.1038/nsmb.1586.
- Hutten, S.; Flotho, A.; Melchior, F.; Kehlenbach, R.H. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Biol Cell 2008, 19, 2300–2310, doi:10.1091/mbc.E07-12-1279.
- Guttler, T.; Gorlich, D. Ran-dependent nuclear export mediators: A structural perspective. EMBO J. 2011, 30, 3457–3474, doi:10.1038/emboj.2011.287.
- Koyama, M.; Matsuura, Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010, 29, 2002–2013, doi:10.1038/emboj.2010.89.
- Culjkovic-Kraljacic, B.; Borden, K.L.B. The Impact of Post-transcriptional Control: Better Living Through RNA Regulons. Genet. 2018, 9, 512, doi:10.3389/fgene.2018.00512.
- Blackinton, J.G.; Keene, J.D. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Cell Dev. Biol. 2014, 34, 44–54, doi:10.1016/j.semcdb.2014.05.014.
- Keene, J.D.; Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005, 13, 327–337.
- Keene, J.D.; Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Cell 2002, 9, 1161–1167.
- Speese, S.D.; Ashley, J.; Jokhi, V.; Nunnari, J.; Barria, R.; Li, Y.; Ataman, B.; Koon, A.; Chang, Y.T.; Li, Q.; et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 2012, 149, 832–846, doi:10.1016/j.cell.2012.03.032.
- Herold, A.; Klymenko, T.; Izaurralde, E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 2001, 7, 1768–1780.
- Wiegand, H.L.; Coburn, G.A.; Zeng, Y.; Kang, Y.; Bogerd, H.P.; Cullen, B.R. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell Biol. 2002, 22, 245–256.
- Carroll, M.; Borden, K.L. The oncogene eIF4E: Using biochemical insights to target cancer. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2013, 33, 227–238.