
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tomasz M. Grzywa | + 1031 word(s) | 1031 | 2020-12-18 06:54:51 |
Video Upload Options
Epithelial–mesenchymal transition (EMT) is a process that is crucial for embryogenesis, wound healing but also malignant progression. EMT leads to the changes in cell–cell and cell–ECM interactions, that allow the migration of epithelial cells and confer them to the mesenchymal phenotype
1. Introduction
Epithelial–mesenchymal transition (EMT) is a process that is crucial for embryogenesis, wound healing but also malignant progression. EMT leads to the changes in cell–cell and cell–ECM interactions, that allow the migration of epithelial cells and confer them to the mesenchymal phenotype [1]. The process can be reversed and it is called a mesenchymal–epithelial transition (MET) and is associated with the colonization of distant organs and the formation of metastases [2].
2. Regulation of Epithelial–Mesenchymal Transition (EMT) by miRNAs
The most important steps are changes in the cell polarity, cytoskeleton and adhesion to other cells and the basement membrane (Figure 1). This allows cells to invade local structures and migrate to distant localizations. Moreover, by undergoing an EMT, carcinoma cells can acquire stem-like cell capabilities such as unlimited self-renewal [3]. EMT is characterized by the downregulation of E-cadherin and the upregulation of N-cadherin and proteases including matrix metalloproteinases such as MMP2 and MMP9 [4]. The most important transcription factors initiating EMT are SNAIL, TWIST, and ZEB. EMT transcription factors regulate the expression of multiple miRNAs [5]. One of the targets of ZEB1 is miR-34a, which regulates multiple properties of tumor cells, including cell migration and invasiveness [5].
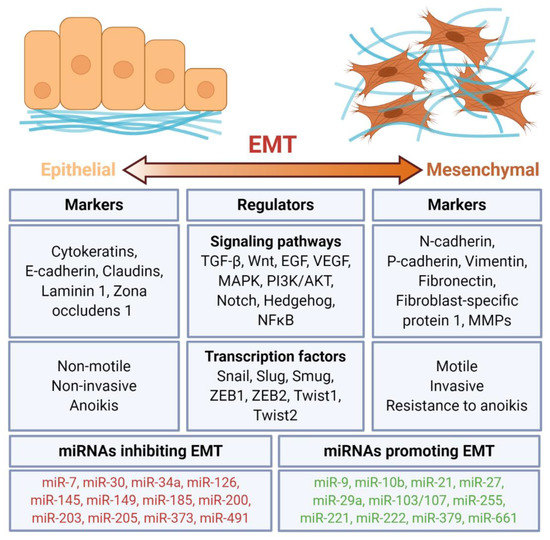
Figure 1. Regulation of epithelial–mesenchymal transition (EMT) by miRNAs. EMT is a process of the acquisition of the mesenchymal features, including motility, invasiveness, and resistance to anoikis of epithelial cells. It is regulated by multiple signaling pathways and transcription factors. Several miRNAs have been identified as either inhibiting or promoting EMT. Created with Biorender.com
All components of EMT signaling pathways are regulated by miRNAs post-transcriptionally [4] (Table 1). In general, EMT is promoted by oncomiRs and inhibited by tumor suppressor miRNAs (Figure 1). A group of oncromiRs called pro-EMT miRNAs promote EMT, tumor cell invasiveness, migration, and metastasis. For example, miR-9 and miR-92a bind to CDH1 which encodes E-cadherin [6]. MiR-10a promotes tumor cell migration and invasion by regulating EMT [7]. In cancer, the maturation of miR-10a is accelerated by XRN2, which leads to increased EMT and metastasis [8]. On the other side, the most important negative regulators of EMT are the miRNA-200 family members. MiR-200s target central regulators of EMT, ZEB1, and ZEB2 [9]. Similarly, miR-22 inhibits EMT via targeting EMT inducer SNAIL and ECM-remodeling MMP14, leading to the suppressed tumor growth, dissemination, and metastasis [10]. MiR-122 triggers a reverse process to EMT, mesenchymal–epithelial transition (MET), and causes cytoskeleton disruption, enhances cell adhesion, and inhibits the migration and invasiveness of cancer cells [11]. A similar effect was exerted by miR-200 family members that induced MET in cancer cell lines [12].The transformation of the growth factor-β (TGF-β) pathway is key signaling inducing EMT. Moreover, TGF-β controls other processes crucial for cancer progression including tumor cell proliferation and invasion. It was shown TGF-β regulates miRNAs expression but is also a target of miRNAs. Several miRNAs were described to be implicated in TGF-β-mediated EMT [13]. Among them, the miR-34 and miR-200 family seem to play the most important role as they form two negative feedback loops with transcription factors involved in EMT. MiR-34 participates with SNAIL1 with the first negative feedback loop and controls the initiation of the EMT process. TGF-β downregulates members of the miR-200 family by the methylation of their promoters and forms an autocrine TGF-β/miR-200b feedback loop. Thus, TGF- β induces EMT by miR-200/ZEB interaction [14]. TGF-β downregulates the expression of miR-584, a negative regulator of PHACTR1 (phosphatase and actin regulator 1), which in turn leads to actin rearrangement and cancer cell migration [15]. Moreover, TGF-β regulates miRNAs targeting adhesion genes [16].
Another crucial signaling pathway for tumor cell invasiveness and metastasis is Wnt/β-catenin. β-catenin-dependent canonical Wnt signaling regulates cell proliferation as well as the development and promotion of EMT, tumor cell invasiveness, and metastasis [17][18]. In the absence of Wnt, β-catenin forms a complex with the tumor suppressor adenomatous polyposis coli gene product (APC), glycogen synthase kinase 3 (GSK3), Axin, and casein kinase 1 (CK1) and is phosphorylated by CK1 and GSK3, which leads to the constant proteasomal degradation of β-catenin. After stimulation with the Wnt ligand, axin is recruited to the membrane complex of the Frizzled (Fz) receptor, low-density lipoprotein receptor-related protein 5 (LRP5) or LRP6 and the scaffolding protein Dishevelled (Dvl), which inhibits the phosphorylation of β-catenin, leading to its stabilization and accumulation in the nucleus [19]. Canonical Wnt signaling is regulated by multiple miRNAs. Wnt1 ligand is targeted by miR-122 [20], miR-148a [21], miR-148b [22], miR-152 [23], miR-329 [24]. Similarly, β-catenin is a target of multiple tumor suppressor miRNAs including miR-33a [25], miR-214 [26], miR-200c [27], miR-320a [28]. WWOX, a β-catenin inhibitor, is targeted by an oncogene, miR-153 [29]. The systemic administration of the miR-153 inhibitor suppressed the development of hepatocellular carcinoma in mice, while tumor cells with upregulated miR-153 expression exhibited increased growth [29]. The expression of miR-374a promotes cell migration and invasiveness by targeting crucial negative regulators of Wnt/β-catenin, including WIF1, PTEN, and WNT5A [30]. Moreover, other components of the Wnt signaling pathway are regulated by miRNAs, including Frizzled-7 by miR-27a [31], LRP6 by miR-202 [32] and miR-432 [33], axin 2 by miR-107 [34], and axin 2 and GSKβ by miR-1246 [35]. Altogether, miRNAs are important regulators of EMT and may either promote or suppress it by targeting different factors (Table 1).
Table 1. Direct regulation of EMT signaling by miRNAs.
|
Target |
miRNAs |
Ref |
|
TGF-β1 |
miR-99a, miR-99b, miR-744 |
|
|
TGFBR2 |
miR-17 family, miR-21, miR-204, miR-211, miR-373, miR-520 |
|
|
ZEB1 |
miR-200 family, miR-205 |
|
|
ZEB2 |
miR-132, miR-138, miR-154, miR-200 family, miR-205, miR-215 |
|
|
Twist1 |
let-7d, miR-33a, miR-145a-5p, miR-151-5p, miR-214, miR-580 |
|
|
Twist2 |
miR-138, miR-215 |
|
|
Notch |
miR-23b, miR-30a, miR-34a, miR-206 |
|
|
Snail1 |
miR-22, miR-34a, miR-137, miR-182 |
|
|
Snail2 |
miR-30a, miR-124, miR-203, miR-204, miR-211 |
|
|
EZH2 |
miR-138 |
[45] |
|
Slug |
miR-1, miR-30a, miR-124, miR-506, miR-630, miR-590-3p |
|
|
N-cadherin |
miR-145, miR-194 |
|
|
Vimentin |
miR-30c |
[72] |
|
Fibronectin |
miR-1, miR-139, miR-200c, miR-432 |
|
|
E-Cadherin |
miR-10b, miR-22, miR-23b, miR-25, miR-92a, miR-221, miR-720, |
|
|
ZO-1 |
miR-103 |
[84] |
|
Claudins |
miR-98 (claudin-1), miR-146-5p (claudin-12), miR-421 (claudin-11), miR-488 (claudin-2), miR-155 (claudin-1) |
References
- Anushka Dongre; Robert A. Weinberg; New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology 2018, 20, 69-84, 10.1038/s41580-018-0080-4.
- Raghu Kalluri; Robert A. Weinberg; The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation 2009, 119, 1420-1428, 10.1172/jci39104.
- Sendurai A. Mani; Wenjun Guo; Mai-Jing Liao; Elinor Ng. Eaton; Ayyakkannu Ayyanan; Alicia Y. Zhou; Mary Brooks; Ferenc Reinhard; Cheng Cheng Zhang; Michail Shipitsin; et al.Lauren L. CampbellKornelia PolyakCathrin BriskenJing YangRobert A. Weinberg The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704-715, 10.1016/j.cell.2008.03.027.
- Samy Lamouille; Jian Xu; Rik Derynck; Molecular mechanisms of epithelial–mesenchymal transition. Nature Reviews Molecular Cell Biology 2014, 15, 178-196, 10.1038/nrm3758.
- Young-Ho Ahn; Don L. Gibbons; Deepavali Chakravarti; Chad J. Creighton; Zain H. Rizvi; Henry P. Adams; Alexander Pertsemlidis; Philip A. Gregory; Josephine A. Wright; Gregory J. Goodall; et al.Elsa R. FloresJonathan M. Kurie ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. Journal of Clinical Investigation 2012, 122, 3170-3183, 10.1172/jci63608.
- Xiang-Ming Ding; MicroRNAs: regulators of cancer metastasis and epithelial-mesenchymal transition (EMT). Chinese Journal of Cancer 2014, 33, 140-147, 10.5732/cjc.013.10094.
- Yan Yan; Qin Wang; Xiao-Ling Yan; Yi Zhang; Wei Li; Fan Tang; Xu Li; Ping Yang; miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition viaEphA8. FEBS Letters 2015, 589, 756-765, 10.1016/j.febslet.2015.02.005.
- H Zhang; Y Lu; E Chen; X Li; B Lv; H G Vikis; P Liu; XRN2 promotes EMT and metastasis through regulating maturation of miR-10a. Oncogene 2017, 36, 3925-3933, 10.1038/onc.2017.39.
- Pui-Wah Choi; Shu-Wing Ng; The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. International Journal of Molecular Sciences 2017, 18, 1207, 10.3390/ijms18061207.
- Q-F Zuo; L-Y Cao; Tianzheng Yu; Leyi Gong; L-N Wang; Y-L Zhao; Bing Xiao; Quan M. Zou; MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death & Disease 2015, 6, e2000-e2000, 10.1038/cddis.2015.297.
- Sheng-Chun Wang; Xiao-Lin Lin; Jing Li; Ting-Ting Zhang; Hui-Yan Wang; Jun-Wen Shi; Sheng Yang; Wen-Tao Zhao; Rao-Ying Xie; Fang Wei; et al.Yu-Juan QinLin ChenJie YangKaitai YaoDong Xiao MicroRNA-122 Triggers Mesenchymal-Epithelial Transition and Suppresses Hepatocellular Carcinoma Cell Motility and Invasion by Targeting RhoA. PLOS ONE 2014, 9, e101330, 10.1371/journal.pone.0101330.
- Golnoush Dehbashi Behbahani; Nastaran Mohammadi Ghahhari; Mohammad Amin Javidi; Asghar Farzi Molan; Neda Feizi; Sadegh Babashah; MicroRNA-Mediated Post-Transcriptional Regulation of Epithelial to Mesenchymal Transition in Cancer. Pathology & Oncology Research 2016, 23, 1-12, 10.1007/s12253-016-0101-6.
- Hiroshi I. Suzuki; MicroRNA Control of TGF-β Signaling. International Journal of Molecular Sciences 2018, 19, 1901, 10.3390/ijms19071901.
- Jingyu Zhang; Xiao-Jun Tian; Hang Zhang; Yue Teng; Ruoyan Li; Fan Bai; Subbiah Elankumaran; Jianhua Xing; TGF- -induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Science Signaling 2014, 7, ra91-ra91, 10.1126/scisignal.2005304.
- Nadège Fils-Aimé; Meiou Dai; Jimin Guo; Mayada El-Mousawi; Bora Kahramangil; Jean-Charles Neel; Jean-Jacques Lebrun; MicroRNA-584 and the Protein Phosphatase and Actin Regulator 1 (PHACTR1), a New Signaling Route through Which Transforming Growth Factor-β Mediates the Migration and Actin Dynamics of Breast Cancer Cells. Journal of Biological Chemistry 2013, 288, 11807-11823, 10.1074/jbc.m112.430934.
- Joanna Bogusławska; Katarzyna Rodzik; Piotr Popławski; Hanna Kędzierska; Beata Rybicka; Elżbieta Sokół; Zbigniew Tański; Agnieszka Piekiełko-Witkowska; TGF-β1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Letters 2018, 412, 155-169, 10.1016/j.canlet.2017.10.019.
- Youn-Sang Jung; Jae-Il Park; Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Experimental & Molecular Medicine 2020, 52, 183-191, 10.1038/s12276-020-0380-6.
- Alanna E. Sedgwick; Crislyn D’Souza-Schorey; Wnt Signaling in Cell Motility and Invasion: Drawing Parallels between Development and Cancer. Cancers 2016, 8, 80, 10.3390/cancers8090080.
- T Zhan; N Rindtorff; Michael Boutros; Wnt signaling in cancer. Oncogene 2016, 36, 1461-1473, 10.1038/onc.2016.304.
- Jun-Gang Zhang; Ying Shi; De-Fei Hong; Mengqi Song; Dongsheng Huang; Chunyou Wang; Gang Zhao; MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/β-catenin pathway. Scientific Reports 2015, 5, srep08087-8087, 10.1038/srep08087.
- Zeinab Ahsani; Samira Mohammadi-Yeganeh; Vahid Kia; Hamzeh Karimkhanloo; Nosratollah Zarghamib; Mahdi Paryan; WNT1 Gene from WNT Signaling Pathway Is a Direct Target of miR-122 in Hepatocellular Carcinoma. Applied Biochemistry and Biotechnology 2016, 181, 884-897, 10.1007/s12010-016-2256-8.
- Han Yan; Xiaogang Dong; Xiaoqin Zhong; Jing Ye; Yun Zhou; Xiaojun Yang; Jian Shen; Jianping Zhang; Inhibitions of epithelial to mesenchymal transition and cancer stem cells‐like properties are involved in miR‐148a‐mediated anti‐metastasis of hepatocellular carcinoma. Molecular Carcinogenesis 2013, 53, 960-969, 10.1002/mc.22064.
- Shifeng Huang; Yan Xie; Ping Yang; Pu Chen; Li-Ping Zhang; HCV Core Protein-Induced Down-Regulation of microRNA-152 Promoted Aberrant Proliferation by Regulating Wnt1 in HepG2 Cells. PLOS ONE 2014, 9, e81730, 10.1371/journal.pone.0081730.
- Liang Wu; Fulai Pei; Xiaojuan Men; Kai Wang; Deliang Ma; miR‑329 inhibits papillary thyroid cancer progression via direct targeting WNT1. Oncology Letters 2018, 16, 3561-3568, 10.3892/ol.2018.9102.
- Yi Fang; YouJun Feng; Tongjin Wu; Swaminath Srinivas; Weiqiang Yang; Jue Fan; Chi Yang; Shihua Wang; Aflatoxin B1 Negatively Regulates Wnt/β-Catenin Signaling Pathway through Activating miR-33a. PLOS ONE 2013, 8, e73004, 10.1371/journal.pone.0073004.
- Xiaojun Wang; Ji Chen; Feng Li; Yanting Lin; Xiaoping Zhang; Zhongwei Lv; Jiaji Jiang; MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of β-catenin. Biochemical and Biophysical Research Communications 2012, 428, 525-531, 10.1016/j.bbrc.2012.10.039.
- Jie Liu; Bai Ruan; Nan You; Qike Huang; Weihui Liu; Zheng Dang; Weihua Xu; Ti Zhou; Ru Ji; Yang Cao; et al.Xia LiDesheng WangKaishan TaoKefeng Dou Downregulation of miR-200a Induces EMT Phenotypes and CSC-like Signatures through Targeting the β-catenin Pathway in Hepatic Oval Cells. PLOS ONE 2013, 8, e79409, 10.1371/journal.pone.0079409.
- Caicheng Lu; Zengwei Liao; Minxian Cai; Guirong Zhang; MicroRNA-320a downregulation mediates human liver cancer cell proliferation through the Wnt/β-catenin signaling pathway. Oncology Letters 2016, 13, 573-578, 10.3892/ol.2016.5479.
- Hong-Wei Hua; Feng Jiang; Qian Huang; Zhijun Liao; Gang Ding; MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget 2015, 6, 3840-3847, 10.18632/oncotarget.2927.
- Junchao Cai; Hongyu Guan; Lishan Fang; Yi Yang; Xun Zhu; Jie Yuan; Jueheng Wu; Mengfeng Li; MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. Journal of Clinical Investigation 2013, 123, 566-579, 10.1172/jci65871.
- Zhaolin Chen; Taotao Ma; Cheng Huang; Lei Zhang; Xiongwen Lv; Tao Xu; Tingting Hu; Jun Li; MiR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/β-catenin pathway in hepatocellular carcinoma cells. Cellular Signalling 2013, 25, 2693-2701, 10.1016/j.cellsig.2013.08.032.
- Yi Zhang; Dayong Zheng; Yan Xiong; Chengbiao Xue; Gen Chen; Bibo Yan; Qifa Ye; miR-202 suppresses cell proliferation in human hepatocellular carcinoma by downregulating LRP6 post-transcriptionally. FEBS Letters 2014, 588, 1913-1920, 10.1016/j.febslet.2014.03.030.
- Nan Jiang; Wen-Jie Chen; Jian-Wen Zhang; Chi Xu; Xian-Cheng Zeng; Tong Zhang; Yang Li; Guo-Ying Wang; Downregulation of miR-432 activates Wnt/β-catenin signaling and promotes human hepatocellular carcinoma proliferation. Oncotarget 2015, 6, 7866-7879, 10.18632/oncotarget.3492.
- Jun-Jie Zhang; Chen-Yu Wang; Long Hua; Kun-Hou Yao; Jiang-Tao Chen; Jun-Hong Hu; miR-107 promotes hepatocellular carcinoma cell proliferation by targeting Axin2. International journal of clinical and experimental pathology 2015, 8, 5168-5174.
- Stella Chai; Kai-Yu Ng; Man Tong; Eunice Y. Lau; Terence K. Lee; Kwok Wah Chan; Yun-Fei Yuan; Tan-To Cheung; Siu-Tim Cheung; Xiao-Qi Wang; et al.Nathalie WongChung-Mau LoKwan ManXin-Yuan GuanStephanie Ma Octamer 4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in liver cancer stem cells. Hepatology 2016, 64, 2062-2076, 10.1002/hep.28821.
- John Martin; Robert L. Jenkins; Rasha Bennagi; Aleksandra Krupa; Aled O. Phillips; Timothy Bowen; Donald J. Fraser; Post-Transcriptional Regulation of Transforming Growth Factor Beta-1 by MicroRNA-744. PLOS ONE 2011, 6, e25044, 10.1371/journal.pone.0025044.
- Gianluca Turcatel; Nicole Rubin; Ahmed El-Hashash; David Warburton; MIR-99a and MIR-99b Modulate TGF-β Induced Epithelial to Mesenchymal Plasticity in Normal Murine Mammary Gland Cells. PLOS ONE 2012, 7, e31032, 10.1371/journal.pone.0031032.
- Sweta Mishra; Janice J. Deng; Pramod S. Gowda; Manjeet K. Rao; Chun-Lin Lin; Chun Liang Chen; Tseshun Huang; Lu-Zhe Sun; Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene 2013, 33, 4097-4106, 10.1038/onc.2013.374.
- Fei E. Wang; Connie Zhang; Arvydas Maminishkis; Lijin Dong; Connie Zhi; Rong Li; Jing Zhao; Vladimir Majerciak; Arti B. Gaur; Shan Chen; et al.Sheldon S. Miller MicroRNA‐204/211 alters epithelial physiology. The FASEB Journal 2010, 24, 1552-1571, 10.1096/fj.08-125856.
- Ali Flores-Pérez; Laurence A. Marchat; Sergio Rodríguez-Cuevas; Verónica Bautista-Piña; Alfredo Hidalgo-Miranda; Elena Aréchaga Ocampo; Mónica Sierra Martínez; Carlos Palma-Flores; Miguel A. Fonseca-Sánchez; Horacio Astudillo-De La Vega; et al.Erika Ruíz-GarcíaJuan Antonio González-BarriosCarlos Pérez-PlasenciaMaría L. StreberCésar López-Camarillo Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Scientific Reports 2016, 6, 34504, 10.1038/srep34504.
- Zeyong Jiang; Jun Yin; Wenfan Fu; Yijun Mo; Youguang Pan; Lu Dai; Haoda Huang; Siwen Li; Jian Zhao; miRNA 17 Family Regulates Cisplatin-Resistant and Metastasis by Targeting TGFbetaR2 in NSCLC. PLOS ONE 2014, 9, e94639, 10.1371/journal.pone.0094639.
- Ioanna Keklikoglou; Cindy Koerner; C Schmidt; J D Zhang; D Heckmann; A Shavinskaya; Heike Allgayer; Brigitte Guckel; Thomas F Fehm; Andreas Schneeweiss; et al.Ozgur SahinStefan WiemannUlrich Tschulena MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 2011, 31, 4150-4163, 10.1038/onc.2011.571.
- Philip A. Gregory; Andrew G. Bert; Emily L. Paterson; Simon C. Barry; Anna Tsykin; Gelareh Farshid; Mathew A. Vadas; Yeesim Khew-Goodall; Gregory J. Goodall; The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature 2008, 10, 593-601, 10.1038/ncb1722.
- Manav Korpal; Esther S. Lee; Guohong Hu; Yibin Kang; The miR-200 Family Inhibits Epithelial-Mesenchymal Transition and Cancer Cell Migration by Direct Targeting of E-cadherin Transcriptional RepressorsZEB1andZEB2. Journal of Biological Chemistry 2008, 283, 14910-14914, 10.1074/jbc.c800074200.
- Xiqiang Liu; Cheng Wang; Zujian Chen; Yi Jin; Yun Wang; Antonia Kolokythas; Yang Dai; Xiaofeng Zhou; MicroRNA-138 suppresses epithelial–mesenchymal transition in squamous cell carcinoma cell lines. Biochemical Journal 2011, 440, 23-31, 10.1042/bj20111006.
- Nicole M A White; Heba W Z Khella; Jorg Grigull; S Adzovic; Yousef M Youssef; R J Honey; Randi Stewart; Kenneth T Pace; G A Bjarnason; Michael A S Jewett; et al.A J EvansManal Y GabrilGeorge M Yousef miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. British Journal of Cancer 2011, 105, 1741-1749, 10.1038/bjc.2011.401.
- Xingyu Lin; Zhiguang Yang; Peng Zhang; Yunpeng Liu; Guoguang Shao; miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncology Letters 2016, 12, 301-306, 10.3892/ol.2016.4577.
- Jiacong You; Yang Li; Nianzhen Fang; Bin Liu; Lingling Zu; Rui Chang; Xuebing Li; Qinghua Zhou; MiR-132 Suppresses the Migration and Invasion of Lung Cancer Cells via Targeting the EMT Regulator ZEB2. PLOS ONE 2014, 9, e91827, 10.1371/journal.pone.0091827.
- Cheng-Chia Yu; Charn-Jung Chang; Chuan-Chih Hsu; Lo-Lin Tsai; Shao-Wei Lu; Hsu-Shan Huang; Jhi-Joung Wang; Ming-Yung Chou; Fang-Wei Hu; Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncology Reports 2011, 26, 1003-1010, 10.3892/or.2011.1360.
- Bin Li; Qingqi Han; Yan Zhu; Yong Yu; Jinghan Wang; Xiaoqing Jiang; Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. The FEBS Journal 2012, 279, 2393-2398, 10.1111/j.1742-4658.2012.08618.x.
- M-L Nairismägi; A Vislovukh; Q Meng; Gueorgui Kratassiouk; C Beldiman; M Petretich; Sonia Groisman; E-M Füchtbauer; Annick Harel-Bellan; I Groisman; et al. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene 2012, 31, 4960-4966, 10.1038/onc.2011.650.
- Maarja-Liisa Nairismägi; Annette Füchtbauer; Rodrigo Labouriau; Jesper Bertram Bramsen; Ernst-Martin Füchtbauer; The Proto-Oncogene TWIST1 Is Regulated by MicroRNAs. PLOS ONE 2013, 8, e66070, 10.1371/journal.pone.0066070.
- Limin Long; Guoqing Huang; Hongyi Zhu; Yonghong Guo; Youshuo Liu; Jirong Huo; Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. Journal of Translational Medicine 2013, 11, 275-275, 10.1186/1479-5876-11-275.
- Jing Zhang; Qi Wang; Zhicheng Quan; Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-β expression. Biochemical and Biophysical Research Communications 2019, 515, 644-650, 10.1016/j.bbrc.2019.05.080.
- Manoela Marques Ortega; Harshita Bhatnagar; An-Ping Lin; Long Wang; Jon C Aster; Heinz Sill; Ricardo C.T. Aguiar; A microRNA-mediated regulatory loop modulates NOTCH and MYC oncogenic signals in B- and T-cell malignancies. Leukemia 2014, 29, 968-976, 10.1038/leu.2014.302.
- Tzu-Ting Huang; Yueh-Hsin Ping; An-Ming Wang; Chia-Chi Ke; Wen-Liang Fang; Kuo-Hung Huang; Hsin-Chen Lee; Chin-Wen Chi; Tien-Shun Yeh; The reciprocal regulation loop of Notch2 pathway and miR-23b in controlling gastric carcinogenesis. Oncotarget 2015, 6, 18012-18026, 10.18632/oncotarget.4000.
- Guisheng Song; Yuxia Zhang; Li Wang; MicroRNA-206 Targetsnotch3, Activates Apoptosis, and Inhibits Tumor Cell Migration and Focus Formation. Journal of Biological Chemistry 2009, 284, 31921-31927, 10.1074/jbc.m109.046862.
- Xuemei Zhang; Feiyan Ai; Xiayu Li; Li Tian; Xiaoyan Wang; Shourong Shen; Fen Liu; MicroRNA-34a suppresses colorectal cancer metastasis by regulating Notch signaling. Oncology Letters 2017, 14, 2325-2333, 10.3892/ol.2017.6444.
- Peixin Dong; Ying Xiong; Hidemichi Watari; Sharon J. B. Hanley; Yosuke Konno; Kei Ihira; Takahiro Yamada; Masataka Kudo; Junming Yue; N. Sakuragi; et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. Journal of Experimental & Clinical Cancer Research 2016, 35, 1-9, 10.1186/s13046-016-0415-y.
- Yun Zhan; Xukun Li; Xiaoshuan Liang; Lin Li; Bangrong Cao; Baona Wang; Jianlin Ma; Fang Ding; Xiang Wang; Da Pang; et al.Zhihua Liu MicroRNA-182 drives colonization and macroscopic metastasis via targeting its suppressor SNAI1 in breast cancer. Oncotarget 2016, 8, 4629-4641, 10.18632/oncotarget.13542.
- Hongping Xia; William K. C. Cheung; Samuel S. Ng; Xiaochun Jiang; Songshan Jiang; Johnny Sze; Gilberto Ka Kit Leung; Gang Lu; Danny T. M. Chan; Xiu-Wu Bian; et al.Hsiang-Fu KungWai Sang PoonMarie C. Lin Loss of Brain-enriched miR-124 MicroRNA Enhances Stem-like Traits and Invasiveness of Glioma Cells. Journal of Biological Chemistry 2012, 287, 9962-9971, 10.1074/jbc.m111.332627.
- Zhiqian Zhang; Baotong Zhang; Weidong Li; Liya Fu; Li Fu; Zhengmao Zhu; Jin-Tang Dong; Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes & Cancer 2011, 2, 782-791, 10.1177/1947601911429743.
- Regalla Kumarswamy; Giridhar Mudduluru; Paolo Ceppi; Santoshi Muppala; Miroslaw Kozlowski; Jacek Niklinski; Mauro Papotti; Heike Allgayer; MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. International Journal of Cancer 2011, 130, 2044-2053, 10.1002/ijc.26218.
- Z Cui; Y Hu; MicroRNA-124 suppresses Slug-mediated lung cancer metastasis.. null 2016, 20, 3802-3811.
- Eiji Tominaga; Katsutoshi Yuasa; Sho Shimazaki; Takao Hijikata; MicroRNA-1 targets Slug and endows lung cancer A549 cells with epithelial and anti-tumorigenic properties. Experimental Cell Research 2013, 319, 77-88, 10.1016/j.yexcr.2012.10.015.
- Chia-Wei Chang; Jyh-Cherng Yu; Yi-Hsien Hsieh; Chung-Chin Yao; Jui-I Chao; Po-Ming Chen; Hsiao-Yen Hsieh; Chia-Ni Hsiung; Hou-Wei Chu; Chen-Yang Shen; et al.Chun-Wen Cheng MicroRNA-30a increases tight junction protein expression to suppress the epithelial-mesenchymal transition and metastasis by targeting Slug in breast cancer. Oncotarget 2016, 7, 16462-16478, 10.18632/oncotarget.7656.
- Wei-Xun Chen; Zhan-Guo Zhang; Ze-Yang Ding; Hui-Fang Liang; Jia Song; Xiao-Long Tan; Jing-Jing Wu; Guang-Zhen Li; Zhuo Zeng; Bi-Xiang Zhang; et al.Xiao-Ping Chen MicroRNA-630 suppresses tumor metastasis through the TGF-β- miR-630-Slug signaling pathway and correlates inversely with poor prognosis in hepatocellular carcinoma. Oncotarget 2016, 7, 22674-22686, 10.18632/oncotarget.8047.
- Meisi Yan; Leiguang Ye; Xinxin Feng; Runze Shi; Zhen Sun; Zhigao Li; Tong Liu; MicroRNA-590-3p inhibits invasion and metastasis in triple-negative breast cancer by targeting Slug.. null 2020, 10, 965-974.
- Han-Bo Ma; Yi-Nan Yao; Jin-Jun Yu; Xue-Xue Chen; Huaifang Li; Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506.. American journal of translational research 2018, 10, 592-604.
- P Gao; A-Y Xing; G-Y Zhou; T-G Zhang; J-P Zhang; C Gao; H Li; D-B Shi; The molecular mechanism of microRNA-145 to suppress invasion–metastasis cascade in gastric cancer. Oncogene 2012, 32, 491-501, 10.1038/onc.2012.61.
- Jinglei Miao; Weiguo Wang; Song Wu; Xiaofang Zang; Yuezhan Li; Jianlong Wang; Ruisen Zhan; Minwei Gao; Minghua Hu; Jinsong Li; et al.Shijie Chen miR-194 Suppresses Proliferation and Migration and Promotes Apoptosis of Osteosarcoma Cells by Targeting CDH2. Cellular Physiology and Biochemistry 2018, 45, 1966-1974, 10.1159/000487973.
- Jessica Bockhorn; Kathy Yee; Ya-Fang Chang; Aleix Prat; Dezheng Huo; Chika Nwachukwu; Rachel Dalton; Simo Huang; Kaitlin E. Swanson; Charles M. Perou; et al.Olufunmilayo I. OlopadeMichael F. ClarkeGeoffrey L. GreeneHuiping Liu MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Research and Treatment 2012, 137, 373-382, 10.1007/s10549-012-2346-4.
- Chuan He Yang; Yinan Wang; Michelle Sims; Chun Cai; Lawrence M. Pfeffer; MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Letters 2019, 465, 59-67, 10.1016/j.canlet.2019.08.021.
- Shanzong Wang; Baohong Gao; Hailin Yang; Xuejian Liu; Xia Wu; Weijuan Wang; MicroRNA‑432 is downregulated in cervical cancer and directly targets FN1 to inhibit cell proliferation and invasion. Oncology Letters 2019, 18, 1475-1482, 10.3892/ol.2019.10403.
- Ying Ye; Juhua Zhuang; Guoyu Wang; Saifei He; Jing Ni; Wei Xia; MicroRNA‑139 targets fibronectin 1 to inhibit papillary thyroid carcinoma progression. Oncology Letters 2017, 14, 7799-7806, 10.3892/ol.2017.7201.
- Hengchun Zhang; Zhiguo Sun; Yan Li; Dong Fan; Hao Jiang; MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomedicine & Pharmacotherapy 2017, 88, 285-292, 10.1016/j.biopha.2017.01.023.
- Carmen Chak-Lui Wong; Aki Pui-Wah Tse; Yan-Ping Huang; Yan-Ting Zhu; David Kung-Chun Chiu; Robin Kit-Ho Lai; Sandy Leung-Kuen Au; Alan Ka-Lun Kai; Joyce Man-Fong Lee; Larry Lai Wei; et al.Felice Ho-Ching TsangRegina Cheuk-Lam LoJue ShiYong-Ping ZhengChun-Ming WongI. O. L. Ng Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology 2014, 60, 1645-1658, 10.1002/hep.27320.
- Swati Dhar; Avinash Kumar; Christian R. Gomez; Israh Akhtar; John C. Hancock; Janice M. Lage; Charles R. Pound; Anait S. Levenson; MTA1-activated Epi-microRNA-22 regulates E-cadherin and prostate cancer invasiveness. FEBS Letters 2017, 591, 924-933, 10.1002/1873-3468.12603.
- Yong Liu; Jing Zhao; Pei-Ying Zhang; Yu Zhang; San-Yuan Sun; Shi-Ying Yu; Qing-Song Xi; MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Medical Science Monitor 2012, 18, BR299-BR308, 10.12659/MSM.883262.
- Yi Pan; Jing Li; Yaqin Zhang; Nan Wang; Hongwei Liang; Yuan Liu; Chen-Yu Zhang; Ke Zen; Hongwei Gu; Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Scientific Reports 2016, 6, 25798, 10.1038/srep25798.
- Jun-Ying Wang; Xiao-Feng Li; Pei-Zhong Li; Xin Zhang; Yu Xu; Xin Jin; MicroRNA-23b regulates nasopharyngeal carcinoma cell proliferation and metastasis by targeting E-cadherin. Molecular Medicine Reports 2016, 14, 537-543, 10.3892/mmr.2016.5206.
- Xiaohui Xu; Zhaoli Chen; Xiaohong Zhao; Jiwen Wang; Dapeng Ding; Zhen Wang; Fengwei Tan; Xiaogang Tan; Fang Zhou; Jian Sun; et al.Nan SunYibo GaoKang ShaoNing LiBin QiuJie He MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochemical and Biophysical Research Communications 2012, 421, 640-645, 10.1016/j.bbrc.2012.03.048.
- Zhao-Li Chen; Xiao-Hong Zhao; Ji-Wen Wang; Bao-Zhong Li; Zhen Wang; Jian Sun; Feng-Wei Tan; Da-Peng Ding; Xiao-Hui Xu; Fang Zhou; et al.Xiao-Gang TanJie HangSu-Sheng ShiXiao-Li FengJie He microRNA-92a Promotes Lymph Node Metastasis of Human Esophageal Squamous Cell Carcinoma via E-Cadherin. Journal of Biological Chemistry 2010, 286, 10725-10734, 10.1074/jbc.m110.165654.
- Jing Du; Fengli Zhang; Ling Zhang; Yueyue Jia; Huixiao Chen; MicroRNA-103 regulates the progression in endometrial carcinoma through ZO-1.. International Journal of Immunopathology and Pharmacology 2019, 33, -, 10.1177/2058738419872621.
- Xianghong Sun; Shichao Cui; Xiaofeng Fu; Chuan Liu; Zhi Wang; Yuanwei Liu; MicroRNA-146-5p promotes proliferation, migration and invasion in lung cancer cells by targeting claudin-12. Cancer Biomarkers 2019, 25, 89-99, 10.3233/cbm-182374.
- Peng Yang; Mei Zhang; Xiting Liu; Huayun Pu; MicroRNA-421 promotes the proliferation and metastasis of gastric cancer cells by targeting claudin-11. Experimental and Therapeutic Medicine 2017, 14, 2625-2632, 10.3892/etm.2017.4798.
- Yong-Bing Wang; Quan Shi; Gang Li; Jun-Hua Zheng; Jie Lin; Wei Qiu; MicroRNA-488 inhibits progression of colorectal cancer via inhibition of the mitogen-activated protein kinase pathway by targeting claudin-2. American Journal of Physiology-Cell Physiology 2019, 316, C33-C47, 10.1152/ajpcell.00047.2018.
- Yi-Feng Zheng; Jie Luo; Guo-Lian Gan; Wei Li; Overexpression of microRNA‐98 inhibits cell proliferation and promotes cell apoptosis via claudin‐1 in human colorectal carcinoma. Journal of Cellular Biochemistry 2018, 120, 6090-6105, 10.1002/jcb.27895.
- Guang-Jun Zhang; Hua-Xu Xiao; Hong-Peng Tian; Zuoliang Liu; Shu-Sen Xia; Tong Zhou; Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. International Journal of Molecular Medicine 2013, 31, 1375-1380, 10.3892/ijmm.2013.1348.




