
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Christopher Collins | -- | 1147 | 2025-07-29 19:51:05 | | | |
| 2 | Catherine Yang | Meta information modification | 1147 | 2025-07-30 02:11:24 | | |
Video Upload Options
Chronic fatigue affects a significant portion of the adult population and presents a diagnostic challenge due to its heterogeneous nature. DNA methylation has emerged as a promising biomarker for systemic physiological states. This study investigates saliva-derived DNA methylation across three CpG sites—cg15957394 (AFAP1), cg13348458 (intergenic on chr6), and cg25294185 (RNASEH2C)—as part of the EpiVitality test developed by Muhdo Health Ltd. Data were obtained from a cohort of 1,260 adults and stratified into two groups: a focus group (N = 571) with daily fatigue scores ≥8, and a complement group (N = 689) with fatigue scores ≤3. Saliva DNA was processed by Eurofins Denmark using Illumina-based analysis. Methylation differences between groups were statistically significant (p = 2.63 × 10⁻¹³, q = 2.63 × 10⁻¹³), with a mean beta difference of -0.028, suggesting an epigenetic link between hypomethylation and fatigue severity.
1. Introduction
Fatigue, particularly chronic fatigue, is a complex symptom influenced by genetic, environmental, and psychological factors. While commonly reported, fatigue is difficult to quantify and differentiate at the molecular level. Recent advances have shown that DNA methylation patterns in accessible tissues such as saliva can reflect biological states including aging, stress, and systemic inflammation. The EpiVitality test targets three CpG loci previously implicated in cellular signalling and genomic stability:
- cg15957394 in the promoter of AFAP1 (Actin Filament Associated Protein 1)
- cg13348458 in an intergenic region on chromosome 6
- cg25294185 in the promoter of RNASEH2C (a subunit of Ribonuclease H2 complex)
This study builds on prior analysis showing hypomethylation with increased stress level [1], extending the methodology to assess fatigue phenotypes using methylation means across these sites which were also used to help identify fibromyalgia [2].
2. Materials and Methods
2.1. Participants and Grouping
Participants (N = 1,260) were divided based on self-reported fatigue scores on a 0–10 scale:
- Focus (Fatigue) Group:
- N = 571
- Fatigue score ≥ 8
- Mean age = 42.3 years (SD = 1.4)
- 42% male
- Complement Group:
- N = 689
- Fatigue score ≤ 3
- Mean age = 41.0 years (SD = 1.2)
- 52% male
2.2. DNA Methylation Profiling
- Saliva samples were collected using standardised collection kits.
- DNA was extracted and processed by Eurofins Denmark using the Illumina platform.
- Beta values were calculated as the ratio of methylated probe intensity to total intensity.
2.3. CpG Sites Analysed
|
Variable Name |
Gene Annotation |
Location |
Function |
|
cg15957394 |
AFAP1 |
chr4:7940563-7941853 |
Promoter |
|
cg13348458 |
Intergenic |
chr6:27569325-27570075 |
Unknown |
|
cg25294185 |
RNASEH2C |
chr11:65487340-65487875 |
Promoter |
2.4 Statistical Analysis
Group means, medians, percentiles, and range distributions were calculated. Significance between groups was assessed using the Mann-Whitney Test.
3. Results
3.1. Group-Wide Methylation Differences
- Mean Methylation (Focus Group): 0.238
- Mean Methylation (Complement Group): 0.266
- Difference in Means: -0.028
- Statistical Significance:
- p = 2.63 × 10⁻¹³
- q = 2.63 × 10⁻¹³
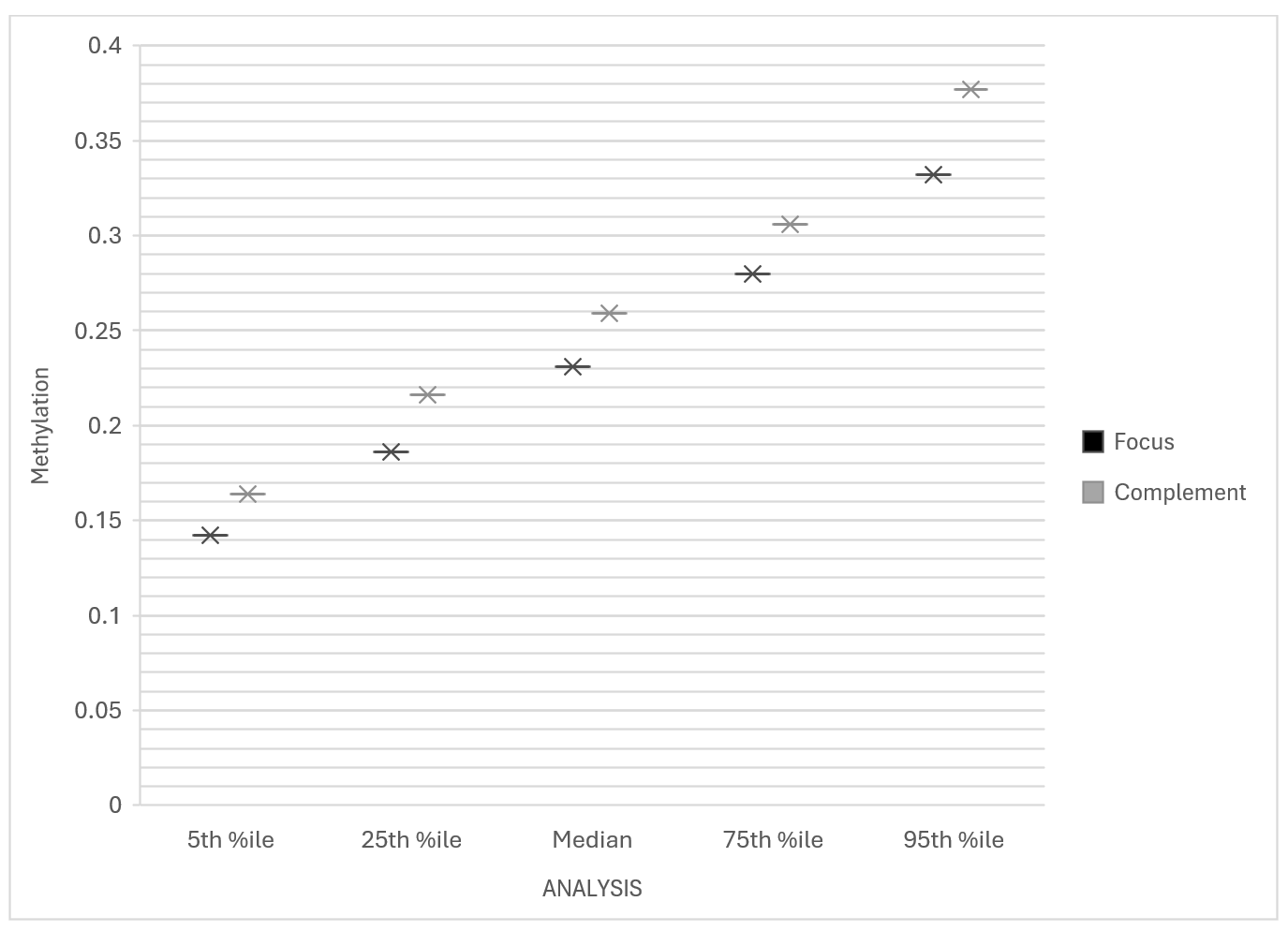
3.2. Distributional Statistics
|
Group |
Min |
5th %ile |
25th %ile |
Median |
75th %ile |
95th %ile |
Max |
|
Focus |
0.094 |
0.142 |
0.186 |
0.231 |
0.280 |
0.332 |
0.553 |
|
Complement |
0.116 |
0.164 |
0.216 |
0.259 |
0.306 |
0.377 |
0.803 |
These values suggest a consistent hypomethylation trend across all three CpG sites in individuals reporting severe fatigue.
4. Discussion
The results demonstrate a robust and statistically significant reduction in methylation at CpG sites within and near AFAP1, RNASEH2C, and an intergenic locus on chr6 among individuals with high fatigue scores:
a. cg15957394 – AFAP1 (Actin Filament Associated Protein 1)
Genomic Location: chr4:7940563–7941853 Region: Promoter
Gene Function:
AFAP1 is involved in actin cytoskeletal remodelling and plays a regulatory role in cell signalling, particularly those involving Src family kinases. It acts as an adaptor protein influencing actin filament integrity, affecting:
Cell motility
Adhesion
Signal transduction pathway
Potential Functional Relevance to Fatigue:
Disruption in cytoskeletal dynamics can impair immune cell trafficking and inflammatory responses, which are tightly linked to fatigue in both acute and chronic conditions. Hypomethylation of the AFAP1 promoter could lead to overexpression, potentially promoting cytoskeletal instability or dysregulated cellular signalling under stress or inflammation.
b. cg13348458 – Intergenic Region on Chromosome 6
Genomic Location: chr6:27569325–27570075 Region: Intergenic
Gene Context:
Although this CpG does not directly map to a known gene, it resides within a region rich in regulatory elements including enhancers and transcription factor binding sites. This locus lies between known immune-related genes such as:
HLA class I and II regions
TRIM family proteins, known to play a role in innate immunity
Functional Relevance to Fatigue:
Intergenic CpG sites often function as long-range enhancers or chromatin regulators. Differential methylation here could reflect systemic immune modulation or neuroinflammatory signalling, both implicated in chronic fatigue syndromes. This CpG may serve as a sentinel site capturing broader epigenetic shifts in immune-regulatory networks.
c. cg25294185 – RNASEH2C (Ribonuclease H2 Subunit C)
Genomic Location: chr11:65487340–65487875 Region: Promoter
Gene Function:
RNASEH2C is part of the Ribonuclease H2 complex, which is essential for:
Removal of ribonucleotides disincorporated into DNA
Maintenance of genome stability
DNA replication and repair processes
Mutations in RNASEH2C have been implicated in Aicardi-Goutières syndrome, an autoimmune encephalopathy, highlighting its role in innate immunity and DNA sensing pathways.
Functional Relevance to Fatigue:
Promoter hypomethylation may lead to altered expression of RNASEH2C, potentially affecting DNA repair efficiency and promoting a persistent inflammatory state via accumulation of nucleic acid debris. This can activate cGAS-STING pathways, triggering fatigue-like symptoms through systemic inflammation.
Importantly, all methylation was measured via saliva DNA, offering a non-invasive, scalable method for epigenetic monitoring. This study supports the potential of the EpiVitality test as a biomarker tool for fatigue, with possible extensions into related conditions such as ME/CFS.
Linking EpiVitality Methylation Results to Chronic Fatigue Syndrome (CFS)
Epigenetic Hypomethylation and Gene Overexpression
The EpiVitality panel revealed significant hypomethylation across three CpG loci in individuals with high fatigue scores. In DNA methylation biology, hypomethylation of promoter regions typically results in increased gene expression. These changes may reflect or contribute to the systemic alterations seen in CFS, which include:
- Aberrant immune activity
- Impaired mitochondrial energy metabolism
- Neuroinflammatory processes
- Genomic instability or DNA damage responses
Hypomethylation of AFAP1’s promoter suggests upregulation of this actin-binding gene. AFAP1 regulates cellular scaffolding and signalling cascades such as Src kinase pathways, which influence. Elevated immune cell activation, cytoskeletal remodelling, and chronic low-grade inflammation are frequently reported in ME/CFS patients. Overactive AFAP1 may contribute to immune cell dysregulation or altered intracellular trafficking, potentially impairing resolution of immune responses and increasing fatigue. This CpG is in a regulatory-rich zone on chromosome 6 (cg13348458), possibly influencing MHC (HLA) gene regions critical for antigen presentation and immune tolerance. Hypomethylation in such regions can lead to dysregulated immune gene expression. CFS has long been associated with immune activation and autoimmune-like profiles, especially following viral triggers like EBV or SARS-CoV-2. Altered methylation in regulatory regions may signal a persistent, maladaptive immune response to prior infections—commonly reported in CFS case histories. RNASEH2C is essential for clearing ribonucleotides from genomic DNA, preventing DNA damage responses and autoimmune triggers. CFS is associated with oxidative stress, mitochondrial dysfunction, and genomic instability. If DNA repair is chronically active, it can lead to activation of DNA sensing pathways (e.g., cGAS-STING) that perpetuate neuroinflammation and sickness behaviour, which includes fatigue, brain fog, and pain.
5. Conclusion
The EpiVitality panel identifies statistically significant methylation differences across three CpG loci in relation to daily fatigue burden. These findings warrant further investigation into epigenetic diagnostics for chronic fatigue syndromes and lifestyle interventions to modulate these markers.
References
- Stress Is Associated with Lowered DNA Methylation. encyclopedia.pub. Retrieved 2025-7-29
- A Test for Fibromyalgia? Saliva DNA Methylation. encyclopedia.pub. Retrieved 2025-7-29




