
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianbo Liu | -- | 3226 | 2024-03-18 09:14:01 | | | |
| 2 | Lindsay Dong | Meta information modification | 3226 | 2024-03-19 02:42:02 | | |
Video Upload Options
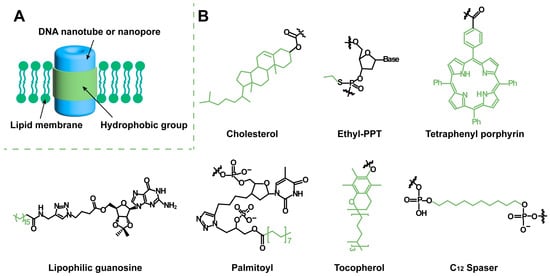
Biomolecular channels on the cell membrane are essential for transporting substances across the membrane to maintain cell physiological activity. Artificial transmembrane channels used to mimic biological membrane channels can regulate intra/extracellular ionic and molecular homeostasis, and they elucidate cellular structures and functionalities. Due to their program design, facile preparation, and high biocompatibility, DNA nanostructures have been widely used as scaffolds for the design of artificial transmembrane channels and exploited for ionic and molecular transport and biomedical applications. DNA-based artificial channels can be designed from two structural modules: DNA nanotubes/nanopores as transport modules for mass transportation and hydrophobic segments as anchor modules for membrane immobilization.
1. Introduction
2. Hydrophobic Modification for Artificial Transmembrane Channels
3. Design of DNA Nanostructure for Artificial Transmembrane Channels
3.1. DNA Wireframe-Based Transmembrane Channels
3.2. DNA Helix Bundle-Based Transmembrane Channels
3.3. DNA Tile-Based Transmembrane Channels
3.4. DNA Origami-Based Transmembrane Channels
3.5. Other DNA-Based Transmembrane Channels
4. Artificial Transmembrane Channels for Biosensing and Biomedical Applications
4.1. DNA-Based Transmembrane Channels for Biosensors
4.1.1. Single-Molecule Nanochannel Sensors
4.1.2. Ligand-Gated Artificial Transmembrane Channels
4.1.3. Environmental Stimuli-Responsive Artificial Transmembrane Channels
4.2. DNA-Based Transmembrane Channels for Biomedical Applications
4.2.1. Cell Mimics for Transmembrane Transport
4.2.2. Transmembrane Channels for Cell Death
References
- Shen, H.; Wang, Y.; Wang, J.; Li, Z.; Yuan, Q. Emerging Biomimetic Applications of DNA Nanotechnology. ACS Appl. Mater. Interfaces 2019, 11, 13859–13873.
- Shen, Q.; Xiong, Q.; Zhou, K.; Feng, Q.; Liu, L.; Tian, T.; Wu, C.; Xiong, Y.; Melia, T.J.; Lusk, C.P.; et al. Functionalized DNA-Origami-Protein Nanopores Generate Large Transmembrane Channels with Programmable Size-Selectivity. J. Am. Chem. Soc. 2022, 145, 1292–1300.
- Jiang, X.; Wang, L.; Liu, S.; Li, F.; Liu, J. Bioinspired artificial nanochannels: Construction and application. Mater. Chem. Front. 2021, 5, 1610–1631.
- Pugh, G.C.; Burns, J.R.; Howorka, S. Comparing proteins and nucleic acids for next-generation biomolecular engineering. Nat. Rev. Chem. 2018, 2, 113–130.
- Luo, Y.; Zhu, C.; Zhang, T.; Yan, T.; Liu, J. Self-assembled Supramolecular Artificial Transmembrane Ion Channels: Recent Progress and Application. Chem. Res. Chin. Univ. 2023, 39, 3–12.
- Langecker, M.; Arnaut, V.; List, J.; Simmel, F.C. DNA nanostructures interacting with lipid bilayer membranes. Acc. Chem. Res. 2014, 47, 1807–1815.
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068.
- Ramezani, H.; Dietz, H. Building machines with DNA molecules. Nat. Rev. Genet. 2020, 21, 5–26.
- Suzuki, Y.; Endo, M.; Sugiyama, H. Mimicking Membrane-Related Biological Events by DNA Origami Nanotechnology. ACS Nano 2015, 9, 3418–3420.
- Wang, D.; Zhang, Y.; Liu, D. DNA nanochannels. F1000Res 2017, 6, 503.
- Niranjan, D.N.; Thiyagarajan, D.; Bhatia, D. DNA Origami in the Quest for Membrane Piercing. Chem. Asian. J. 2022, 17, e202200591.
- Chen, J.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633.
- Ohmann, A.; Li, C.Y.; Maffeo, C.; Nahas, K.A.; Baumann, K.N.; Gopfrich, K.; Yoo, J.; Keyser, U.F.; Aksimentiev, A. A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nat. Commun. 2018, 9, 2426.
- Jones, S.F.; Joshi, H.; Terry, S.J.; Burns, J.R.; Aksimentiev, A.; Eggert, U.S.; Howorka, S. Hydrophobic Interactions between DNA Duplexes and Synthetic and Biological Membranes. J. Am. Chem. Soc. 2021, 143, 8305–8313.
- Chidchob, P.; Offenbartl-Stiegert, D.; McCarthy, D.; Luo, X.; Li, J.; Howorka, S.; Sleiman, H.F. Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions. J. Am. Chem. Soc. 2018, 141, 1100–1108.
- Burns, J.R.; Howorka, S. Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy. ACS Nano 2018, 12, 3263–3271.
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936.
- Gopfrich, K.; Li, C.Y.; Ricci, M.; Bhamidimarri, S.P.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10, 8207–8214.
- Diederichs, T.; Pugh, G.; Dorey, A.; Xing, Y.; Burns, J.R.; Nguyen, Q.H.; Tornow, M.; Tampe, R.; Howorka, S. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 2019, 10, 5018.
- Thomsen, R.P.; Malle, M.G.; Okholm, A.H.; Krishnan, S.; Bohr, S.S.; Sorensen, R.S.; Ries, O.; Vogel, S.; Simmel, F.C.; Hatzakis, N.S.; et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 2019, 10, 5655.
- Fragasso, A.; De Franceschi, N.; Stommer, P.; van der Sluis, E.O.; Dietz, H.; Dekker, C. Reconstitution of Ultrawide DNA Origami Pores in Liposomes for Transmembrane Transport of Macromolecules. ACS Nano 2021, 15, 12768–12779.
- Xing, Y.; Dorey, A.; Jayasinghe, L.; Howorka, S. Highly shape- and size-tunable membrane nanopores made with DNA. Nat. Nanotechnol. 2022, 17, 708–713.
- Burns, J.R.; Stulz, E.; Howorka, S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013, 13, 2351–2356.
- Burns, J.R.; Gopfrich, K.; Wood, J.W.; Thacker, V.V.; Stulz, E.; Keyser, U.F.; Howorka, S. Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angew. Chem. Int. Ed. Engl. 2013, 52, 12069–12072.
- Debnath, M.; Chakraborty, S.; Kumar, Y.P.; Chaudhuri, R.; Jana, B.; Dash, J. Ionophore constructed from non-covalent assembly of a G-quadruplex and liponucleoside transports K+-ion across biological membranes. Nat. Commun. 2020, 11, 469.
- Krishnan, S.; Ziegler, D.; Dietz, H.; Simmel, F.C. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016, 7, 12787.
- Li, C.; Chen, H.; Zhou, L.; Shi, H.; He, X.; Yang, X.; Wang, K.; Liu, J. Single-stranded DNA designed lipophilic G-quadruplexes as transmembrane channels for switchable potassium transport. Chem. Commun. 2019, 55, 12004–12007.
- Aldaye, F.A.; Lo, P.K.; Karam, P.; McLaughlin, C.K.; Cosa, G.; Sleiman, H.F. Modular construction of DNA nanotubes of tunable geometry and single- or double-stranded character. Nat. Nanotechnol. 2009, 4, 349–352.
- Lo, P.K.; Karam, P.; Aldaye, F.A.; McLaughlin, C.K.; Hamblin, G.D.; Cosa, G.; Sleiman, H.F. Loading and selective release of cargo in DNA nanotubes with longitudinal variation. Nat. Chem. 2010, 2, 319–328.
- Hamblin, G.D.; Carneiro, K.M.M.; Fakhoury, J.F.; Bujold, K.E.; Sleiman, H.F. Rolling Circle Amplification-Templated DNA Nanotubes Show Increased Stability and Cell Penetration Ability. J. Am. Chem. Soc. 2012, 134, 2888–2891.
- Hariri, A.A.; Hamblin, G.D.; Gidi, Y.; Sleiman, H.F.; Cosa, G. Stepwise growth of surface-grafted DNA nanotubes visualized at the single-molecule level. Nat. Chem. 2015, 7, 295–300.
- Rahbani, J.F.; Vengut-Climent, E.; Chidchob, P.; Gidi, Y.; Trinh, T.; Cosa, G.; Sleiman, H.F. DNA Nanotubes with Hydrophobic Environments: Toward New Platforms for Guest Encapsulation and Cellular Delivery. Adv. Health Mater. 2018, 7, 1701049.
- Saliba, D.; Luo, X.; Rizzuto, F.J.; Sleiman, H.F. Programming rigidity into size-defined wireframe DNA nanotubes. Nanoscale 2023, 15, 5403–5413.
- Yoo, J.; Aksimentiev, A. Molecular Dynamics of Membrane-Spanning DNA Channels: Conductance Mechanism, Electro-Osmotic Transport, and Mechanical Gating. J. Phys. Chem. Lett. 2015, 6, 4680–4687.
- Maingi, V.; Lelimousin, M.; Howorka, S.; Sansom, M.S.P. Gating-like Motions and Wall Porosity in a DNA Nanopore Scaffold Revealed by Molecular Simulations. ACS Nano 2015, 9, 11209–11217.
- Maingi, V.; Burns, J.R.; Uusitalo, J.J.; Howorka, S.; Marrink, S.J.; Sansom, M.S. Stability and dynamics of membrane-spanning DNA nanopores. Nat. Commun. 2017, 8, 14784.
- Birkholz, O.; Burns, J.R.; Richter, C.P.; Psathaki, O.E.; Howorka, S.; Piehler, J. Multi-functional DNA nanostructures that puncture and remodel lipid membranes into hybrid materials. Nat. Commun. 2018, 9, 1521.
- Mathieu, F.; Liao, S.; Kopatsch, J.; Wang, T.; Mao, C.; Seeman, N.C. Six-Helix Bundles Designed from DNA. Nano Lett. 2005, 5, 661–665.
- Endo, M.; Seeman, N.C.; Majima, T. DNA Tube Structures Controlled by a Four-Way-Branched DNA Connector. Angew. Chem. Int. Ed. Engl. 2005, 44, 6074–6077.
- Mohammed, A.M.; Šulc, P.; Zenk, J.; Schulman, R. Self-assembling DNA nanotubes to connect molecular landmarks. Nat. Nanotechnol. 2016, 12, 312–316.
- Green, L.N.; Subramanian, H.K.K.; Mardanlou, V.; Kim, J.; Hariadi, R.F.; Franco, E. Autonomous dynamic control of DNA nanostructure self-assembly. Nat. Chem. 2019, 11, 510–520.
- Agarwal, S.; Franco, E. Enzyme-Driven Assembly and Disassembly of Hybrid DNA-RNA Nanotubes. J. Am. Chem. Soc. 2019, 141, 7831–7841.
- Jia, S.; Phua, S.C.; Nihongaki, Y.; Li, Y.; Pacella, M.; Li, Y.; Mohammed, A.M.; Sun, S.; Inoue, T.; Schulman, R. Growth and site-specific organization of micron-scale biomolecular devices on living mammalian cells. Nat. Commun. 2021, 12, 5729.
- Dhanasekar, N.N.; Li, Y.; Schulman, R. The ion permeability of DNA nanotube channels. BioRxiv 2022, 1–33.
- Li, Y.; Maffeo, C.; Schulman, R. Leakless end-to-end transport of small molecules through micron-length DNA nanochannels. Sci. Adv. 2022, 8, eabq4834.
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302.
- Forman, S.L.; Fettinger, J.C.; Pieraccini, S.; Gottarelli, G.; Davis, J.T. Toward Artificial Ion Channels: A Lipophilic G-Quadruplex. J. Am. Chem. Soc. 2000, 122, 4060–4067.
- Lee, M.P.H.; Parkinson, G.N.; Hazel, P.; Neidle, S. Observation of the Coexistence of Sodium and Calcium Ions in a DNA G-Quadruplex Ion Channel. J. Am. Chem. Soc. 2007, 129, 10106–10107.
- Akhshi, P.; Mosey, N.J.; Wu, G. Free-Energy Landscapes of Ion Movement through a G-Quadruplex DNA Channel. Angew. Chem. 2012, 124, 2904–2908.
- Akhshi, P.; Wu, G. Umbrella sampling molecular dynamics simulations reveal concerted ion movement through G-quadruplex DNA channels. Phys. Chem. Chem. Phys. 2017, 19, 11017–11025.
- Balasubramanian, S.; Senapati, S. Dynamics and Barrier of Movements of Sodium and Potassium Ions Across the Oxytricha nova G-Quadruplex Core. J. Phys. Chem. B 2020, 124, 11055–11066.
- Seifert, A.; Gopfrich, K.; Keyser, U.F.; Howorka, S. Bilayer-Spanning DNA Nanopores with Voltage-Switching between Open and Closed State. ACS Nano 2015, 9, 1117–1126.
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156.
- Dey, S.; Dorey, A.; Abraham, L.; Xing, Y.; Zhang, I.; Zhang, F.; Howorka, S.; Yan, H. A reversibly gated protein-transporting membrane channel made of DNA. Nat. Commun. 2022, 13, 2271.
- Arnott, P.M.; Howorka, S. A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport across Membranes. ACS Nano 2019, 13, 3334–3340.
- Li, P.; Xie, G.; Kong, X.Y.; Zhang, Z.; Xiao, K.; Wen, L.; Jiang, L. Light-Controlled Ion Transport through Biomimetic DNA-Based Channels. Angew. Chem. Int. Ed. Engl. 2016, 55, 15637–15641.
- Offenbartl-Stiegert, D.; Rottensteiner, A.; Dorey, A.; Howorka, S. A Light-Triggered Synthetic Nanopore for Controlling Molecular Transport Across Biological Membranes. Angew. Chem. Int. Ed. Engl. 2022, 61, e202210886.
- Li, C.; Chen, H.; Yang, X.; Wang, K.; Liu, J. An ion transport switch based on light-responsive conformation-dependent G-quadruplex transmembrane channels. Chem. Commun. 2021, 57, 8214–8217.
- Li, C.; Chen, H.; Chen, Q.; Shi, H.; Yang, X.; Wang, K.; Liu, J. Lipophilic G-Quadruplex Isomers as Biomimetic Ion Channels for Conformation-Dependent Selective Transmembrane Transport. Anal. Chem. 2020, 92, 10169–10176.
- Messager, L.; Burns, J.R.; Kim, J.; Cecchin, D.; Hindley, J.; Pyne, A.L.; Gaitzsch, J.; Battaglia, G.; Howorka, S. Biomimetic Hybrid Nanocontainers with Selective Permeability. Angew. Chem. Int. Ed. Engl. 2016, 55, 11106–11109.
- Burns, J.R.; Al-Juffali, N.; Janes, S.M.; Howorka, S. Membrane-spanning DNA nanopores with cytotoxic effect. Angew. Chem. Int. Ed. Engl. 2014, 53, 12466–12470.
- Lv, C.; Gu, X.; Li, H.; Zhao, Y.; Yang, D.; Yu, W.; Han, D.; Li, J.; Tan, W. Molecular Transport through a Biomimetic DNA Channel on Live Cell Membranes. ACS Nano 2020, 14, 14616–14626.




