Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Evangelos Zoidis | -- | 3606 | 2024-03-15 09:57:00 | | | |
| 2 | Jessie Wu | + 4 word(s) | 3610 | 2024-04-15 10:01:32 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kouvedaki, I.; Pappas, A.C.; Surai, P.F.; Zoidis, E. Oxidative Stress and Poultry. Encyclopedia. Available online: https://encyclopedia.pub/entry/56333 (accessed on 07 February 2026).
Kouvedaki I, Pappas AC, Surai PF, Zoidis E. Oxidative Stress and Poultry. Encyclopedia. Available at: https://encyclopedia.pub/entry/56333. Accessed February 07, 2026.
Kouvedaki, Ioanna, Athanasios C. Pappas, Peter F. Surai, Evangelos Zoidis. "Oxidative Stress and Poultry" Encyclopedia, https://encyclopedia.pub/entry/56333 (accessed February 07, 2026).
Kouvedaki, I., Pappas, A.C., Surai, P.F., & Zoidis, E. (2024, March 15). Oxidative Stress and Poultry. In Encyclopedia. https://encyclopedia.pub/entry/56333
Kouvedaki, Ioanna, et al. "Oxidative Stress and Poultry." Encyclopedia. Web. 15 March, 2024.
Copy Citation
A challenge facing the poultry industry is related to the spread of pathogens within commercial farms and, consequently, its high dependence on antibiotics and other pharmaceuticals. Although the inclusion of antibiotics at sub-therapeutical levels in broiler diets has proven to be an efficient strategy through which to suppress the pathogenic bacteria in the gut and enhance animal performance, their usage as growth promoters has been banned in Europe due to concerns regarding the consequences of antibiotic resistance on human health. Under this context, plenty of phytochemicals and antioxidants are being explored in broiler diets.
broilers
gene expression
natural antioxidants
oxidative stress
redox homeostasis
Nrf2
1. Introduction
The challenge of feeding 9 billion people, by 2050, will determine the future of humanity [1]. Taking into account that the global population will continue to grow, the world is now facing a new challenge: food security. There is a competition for food resources on the ground, where a large percentage of humans, unfortunately, still do not have access to sufficient dietary protein and energy, let alone those that suffer from some form of micronutrient malnourishment [2]. Under this scope, there is a pressing need to tackle food security challenges under a dynamic multidimensional management of food–feed competition and environmental sustainability where animal protein is concerned. Worldwide, the leading meat production sector is the poultry industry as it efficiently supplies high-quality animal protein to the world. The meat poultry industry is dominated by the chicken meat industry, which is also known as the broiler industry [3]. At a global scale, the poultry sector is expected to continuously grow at a compound annual growth rate (CAGR) of 6.8% by 2030 [4].
Broilers, as is the case for all aerobic living organisms, are capable of generating ROS (reactive oxygen species) to regulate important physiological functions while keeping them at low levels to maintain redox balance. However, there are many unfavorable conditions and challenges, either of endogenous (related to broiler) or exogenous (related to farm conditions) origin, which can disturb this balance through the overproduction of ROS at levels that the endogenous antioxidant system cannot cope with. As a result, lipids, proteins, DNA, and carbohydrates may be damaged, thereby inducing the organism’s inability to effectively absorb essential nutrients and thus leading to deteriorating conditions. Supplementation with natural antioxidants can also enhance its function by affecting gene expression and regulation, as well as apoptosis and signal transduction [5].
2. The Endogenous Antioxidant System
In recent decades, it has become apparent that the excessive production of free radicals resulting in oxidative stress and redox imbalance constitutes the primary causative factors for the detrimental consequences of stress in poultry. Oxidative stress is characterized by an imbalance between oxidants and antioxidants. It can be further delineated into ‘oxidative eustress’ at low levels and ‘oxidative distress’ at high levels. At low levels, ROS, which is generated as the by-products of aerobic metabolism, interact with specific targets and play a significant role in redox signaling. They are responsible for stress adaptation, homeostasis, and the maintenance of health. However, exposure to high levels of ROS can lead to biomolecule damage, thus causing oxidative distress—which, in turn, disrupts redox signaling. This disruption can have a profound impact on the productive and reproductive performance of poultry. Throughout the course of evolution, integrated antioxidant defense systems were developed in poultry. These defense mechanisms are designed to regulate the production of free radicals and uphold the balance between antioxidants and prooxidants in the redox environment [5][6][7].
It has been suggested that the endogenous antioxidant system operates on three principal levels, which comprise natural fat-soluble antioxidants such as vitamin E, carotenoids, and ubiquinones; water-soluble antioxidants like ascorbic acid, uric acid, carnitine, and taurine; and a collection of antioxidant enzymes, namely glutathione peroxidase (GPx), catalase (CAT), and superoxide dismutase (SOD). Additionally, the thiol redox system—which encompasses the glutathione system (comprising glutathione, glutathione reductase, glutaredoxin, and glutathione peroxidase) and the thioredoxin system (comprising thioredoxin, thioredoxin reductase, thioredoxin peroxidase such as peroxiredoxins, and sulfiredoxin)—is integral to this comprehensive antioxidant defense mechanism [5].
The first level of defense, which is tasked with averting radical formation, sustains the redox balance, as well as participates in cell signaling (which encompasses essential enzymes like SOD, GPx, CAT, and metal-binding proteins, but also the thioredoxin system, glutathione system, vitagenes, and transcription factors such as Nrf2 (nuclear factor erythroid 2-related factor 2), NF-κB (nuclear factor kappa B), and HSF (heat shock factor)). Since the superoxide radical (O2•) is the primary radical produced under normal cellular conditions, SOD is regarded as a key component in the first level of defense. Through its involvement, O2• is transformed into hydroperoxide (H2O2). Recognized as toxic to cells, H2O2 undergoes a detoxification that is facilitated by a range of enzymes, including catalase, GPx, and peroxiredoxins [5].
Despite its best efforts, the first level of defense proves insufficient in completely preventing the formation of free radicals. This inadequacy results in the release of radicals that are capable of initiating lipid peroxidation and causing damage to DNA and proteins. Consequently, a second level of defense was established, which comprises chain-breaking antioxidants such as vitamin E, ubiquinol, carotenoids, ascorbic acid, uric acid, carnitine, and taurine, as well as various other antioxidants. These compounds work to inhibit peroxidation by minimizing the chain length of the propagation reaction. Vitamin E is recognized as the most effective natural free radical scavenger; however, it can lead to the production of hydroperoxides that are toxic to the cell. Thus, vitamin E and GPx operate collaboratively, thereby offering a comprehensive and effective antioxidant defense. Coenzyme Q, also recognized as ubiquinone, actively participates in shielding biological molecules, including lipids, proteins, and DNA, from oxidative damage. It accomplishes this by neutralizing free radicals, replenishing other antioxidants (such as vitamins E and C), and overseeing the maintenance of mitochondrial integrity. Additionally, the second level of antioxidant defense involves the active participation of the glutathione and thioredoxin systems. Glutathione (GSH), an abundant non-protein thiol in avian cells, stands as one of the most crucial non-enzymatic antioxidants in poultry. It actively contributes to the maintenance of redox balance and signaling, as well as to the regulation of transcription factors and gene expression [5].
Nevertheless, even the second level of antioxidant defense proves inadequate in preventing the detrimental impact of reactive oxygen species (ROS) on lipids, proteins, and DNA. In such instances, the third level of defense relies on systems that are designed to either eliminate or repair damaged molecules. This level encompasses lipolytic enzymes (lipases) and proteolytic enzymes (peptidases or proteases), as well as various other enzymes (DNA repair enzymes, ligases, nucleases, polymerases, proteinases, phospholipases, various transferases, etc.). Additionally, protein chaperones, including heat shock proteins (HSPs), play a crucial role in this third level of defense [5].
Antioxidants function collectively within the body, and they constitute an integrated system that is designed to mitigate oxidative stress. However, these adaptive mechanisms exhibit limited efficacy. Once the production of free radicals surpasses the antioxidant system’s capacity for neutralization, lipid peroxidation ensues, thus resulting in damage to unsaturated lipids in cell membranes, amino acids in proteins, and nucleotides in DNA. Consequently, membrane and cell integrity become compromised [5].
3. Oxidative Stress and Gene Expression
Under oxidative stress, the body initiates a response process that includes the activation of the gene expression of the defense systems. Mitogen-activated protein kinases (MAPK) are important in the cell’s response to the hormones, cytokines, and signals generated by oxidative stress. Apoptosis signal-regulating kinase 1 (ASK1), which is activated in various stressful situations, causes the upregulation of MAPK. Several proteins appear to influence the action of ASK1. For example, thioredoxin can lead to its direct inhibition as its reduced form can bind to ASK1, thereby inhibiting its dimerization and activation. During oxidative stress, thioredoxin is oxidized, which results in its uncoupling from ASK1, thus enabling its activation and involvement in the p38 signal transduction and leading to non-apoptotic effects. ROS can either stimulate protein kinases G (PKG), -A (PKA), and -C (PKC), which—in turn—can regulate MAPK activation or have an immediate impact on activating the MAPK pathway as they can act directly on MAPK phosphatases (MKPs). The MAPK subfamilies include kinases such as ERK (extracellular regulated kinases, i.e., ERK1/2, ERK3/4, ERK5, and ERK7/ERK8); p38; JNK (c-Jun N-terminal kinase); and NLK (Nemo-like kinase) [8][9]. The ERK pathway is mainly associated with regulating cell proliferation, while that of p38 and JNK is related to stress. For this reason, the latter two are grouped together and referred to as stress-activated protein kinases (SAPK) [10]. The most important effect of ROS on MAPKs involves the regulation of transcription factors that control the expression of many protective genes, which contributes to the termination of impaired cell division and leads to apoptosis [11]. Such factors are Nrf2 (nuclear factor erythroid 2-related factor 2) and NF-κB (nuclear factor kappa B). The transcription factor Nrf2 is responsible for the activation of genes that lead to the synthesis of protective antioxidant molecules, while NF-κB activates the genes involved in inflammatory, immune, and acute phase responses. Oxidative stress can cause damage to various molecules in the body. In response to this, other reactions, such as HSR (heat shock response), UPR (unfolded protein response), HIR (hypoxia-induced response), DNA damage response, etc., are triggered to regulate the corresponding transcription factors like HSF1 (heat shock factor 1), FOXO (forkhead box O), HIF (hypoxia-inducible factor), p53 (cellular tumor antigen 53), etc. (as shown in Figure 1).
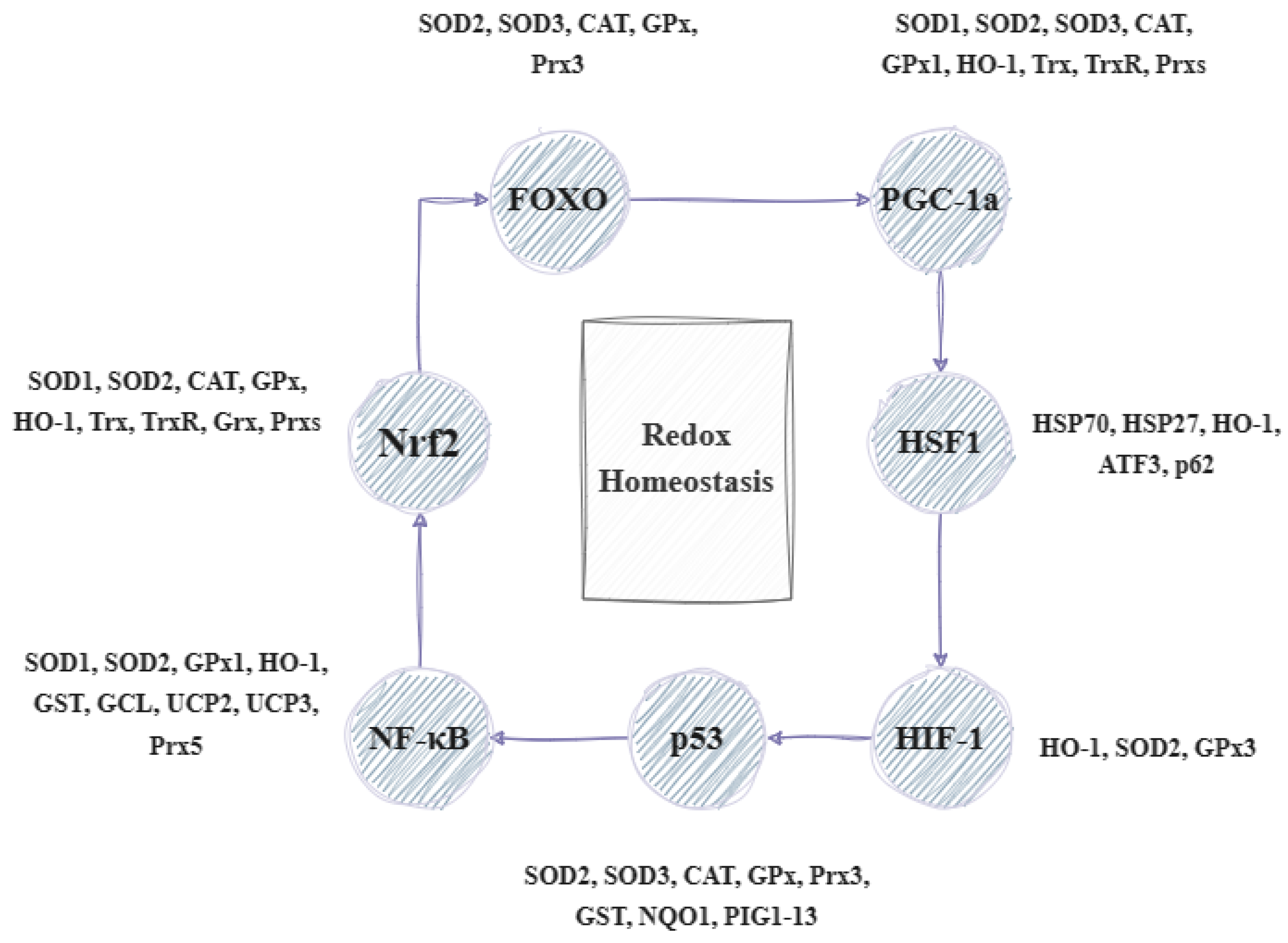
Figure 1. Illustration of the transcriptional factors and genes involved in the regulation of redox homeostasis (based on [5] with modifications). FOXO: forkhead box O transcription factor, PGC-1a: peroxisome proliferator-activated receptor-gamma coactivator, HSF1: heat shock factor 1; HIF-1: hypoxia-inducible factor 1; p53: cellular tumor antigen 53; NF-kB: nuclear factor kappa B; Nrf2: nuclear respiratory factor 2; SOD1–3: superoxide dismutase 1–3; CAT: catalase; GPx: glutathione peroxidase; Prx3,5: peroxiredoxin 3, 5; HO-1: heme oxygenase-1; Trx: thioredoxin; TrxR: thioredoxin reductase; Grx: glutaredoxin; GST: glutathione S-transferase; GCL: glutamate–cysteine ligase; UCP2,3: uncoupling protein 1, 3; NQO1: NAD(P)H:quinone oxidoreductase 1; PIG1–13: p53-inducible genes 1–13; and ATF3: activating transcription factor 3.
These factors lead to the regulation of several genes that help in stress sensing and maintaining redox homeostasis. These genes are known as vitagenes and were first described in 1998 to signify the body’s efficient functioning in repairing and maintaining itself through a variety of processes regulated by these genes. The term ‘vitagenes’ was later coined by the medical sciences to refer to a group of genes that are responsible for upholding cellular homeostasis during periods of stress. This group encompasses several gene families such as heat shock proteins (HSPs), the thioredoxin system, sirtuins, and SOD (superoxide dismutase). HSPs comprise HO-1 (heme oxygenase-1) and HSP70 (heat shock protein 70), while SODs include SOD1, SOD2, and SOD3. The thioredoxin and glutathione system involves Trx (thioredoxin), TrxR (thioredoxin reductase), Prx (peroxiredoxin), Srx (sulfiredoxin), and GSH (glutathione), as well as GR (glutathione reductase), GPx (glutathione peroxidase), and Grx (glutaredoxin) [12]. Lastly, sirtuins (SIRTs) consist of SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7. The products of these genes can detect various forms of stress and regulate the production of different protective molecules. Sirtuins can influence the expression and activity of many factors. For instance, SIRT1 can combat oxidative stress by regulating transcription factors, such as PPAR (peroxisome proliferator-activated receptor), NRF (nuclear respiratory factor), and TFAM (mitochondrial transcription factor A). Moreover, it aids in enhancing the expression of SOD and GPX while also triggering the FOXO/MnSOD signaling pathway by regulating the acetylation of the FOXO factor. This ultimately results in an increase in the expression of MnSOD and catalase, which can effectively counteract oxidative stress. It can also regulate the inflammatory response by acting on NF-κB and thus affecting the transcription of genes such as IL-1 (interleukin-1), TNF-α (tumor necrosis factor α), IL-8 (interleukin-8), and IL-6 (interleukin-6). Similarly, vitagenes can be affected by various transcription factors. The activation of factors such as HSF and FOXO results in the increased production of HSP70 and sirtuins. Similarly, the activation of Nrf2 leads to an increased synthesis of SOD, HO-1, and the compounds in the glutathione and thioredoxin systems. Certain vitagenes can be activated without the involvement of transcription factors. When the body’s antioxidant defense system is unable to forestall the damage caused by ROS, a range of mechanisms, including mitophagy, autophagy, apoptosis, necroptosis, and ferroptosis, are implemented (Figure 2) [5][6][13][14].
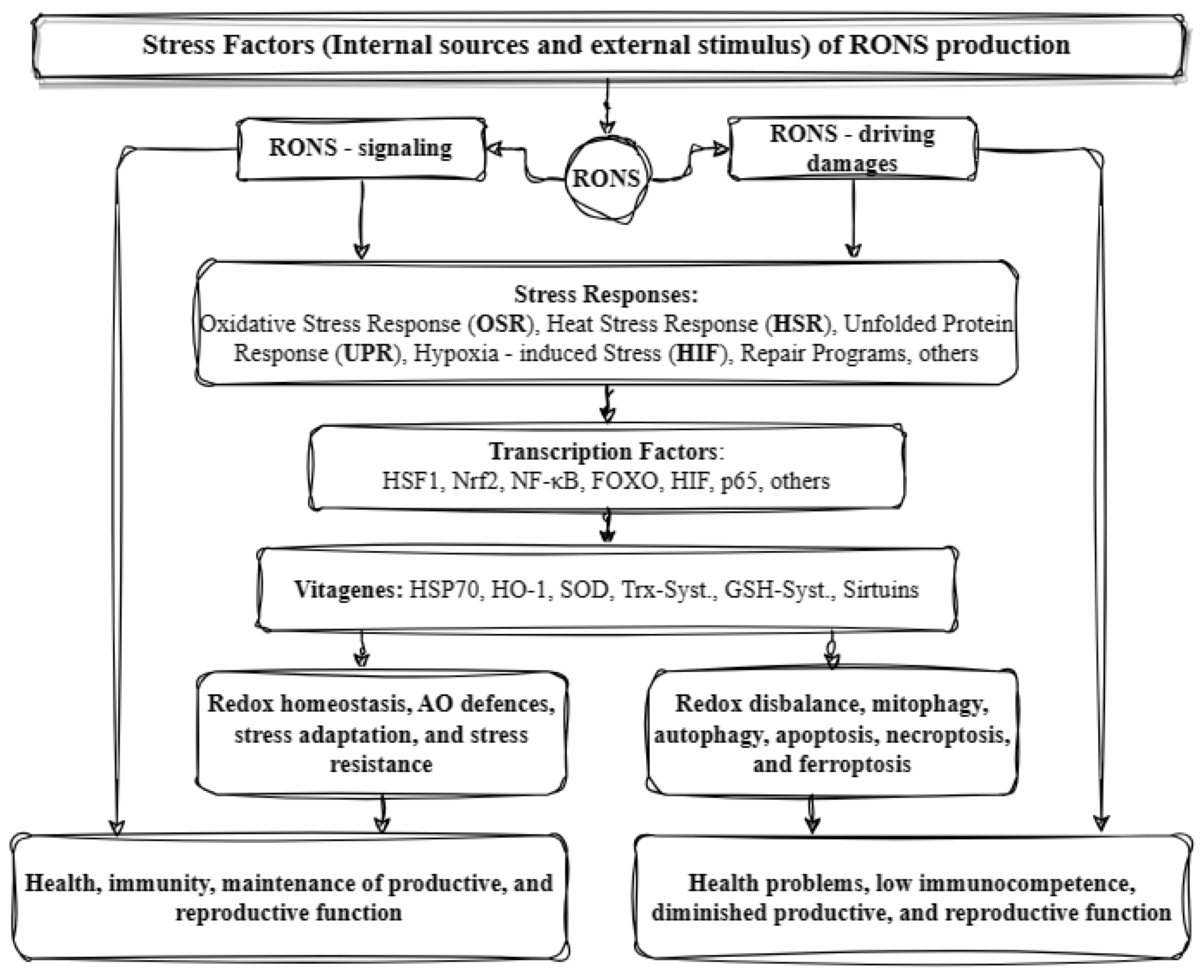
Figure 2. Illustration of the antioxidant defense system in animals (based on [6] with modifications). RONS: reactive oxygen and nitrogen species; HSF1: heat shock factor 1; Nrf2: nuclear respiratory factor 2; NF-kB: nuclear factor kappa B; FOXO: forkhead box O transcription factor; HIF: hypoxia-inducible factor; p65: cellular tumor antigen 65; HSPs: heat shock proteins, HO-1: heme oxygenase 1; SOD: superoxide dismutase; Trx-Syst.: thioredoxin system; GSH-Syst.: glutathione system; and AO: antioxidant.
The process of apoptosis through the mitochondrial pathway is regulated by a group of proteins known as the B-cell lymphoma 2 (Bcl-2) family. With over 30 proteins in this family, each contain up to four conserved Bcl-2 homology domains (BH1–4) that are essential for their function, with BH3 being the death domain. This family of proteins can be divided into two categories: anti-apoptotic survival proteins and pro-apoptotic proteins. The anti-apoptotic proteins include Bcl-2, Bcl-XL, Bcl-W, Bcl-B, A1, and Mcl-1. Conversely, the pre-apoptotic proteins include Bax, Bak, Bok, Bok, Bad, Bik, Bmf, Hrk, SOUL, Bid, Bim, Puma, and Noxa. Each cell comprises an assortment of proteins from both categories, thereby indicating their interdependence and functional significance. The regulation of these proteins is crucial for cell survival. Apoptosis can be triggered by the activation of TNF-R (tumor necrosis factor receptors). Studies have shown that SIRT1 may impact the transcription of genes such as IAP (the inhibitor of apoptosis protein), Bcl-2, and TNF-R. Autophagy, on the other hand, is regulated by autophagy-related genes (Atgs) and the kinase pathway known as the mammalian target of rapamycin (mTOR). Autophagy, a process that leads to the degradation of cellular components, is inhibited by the mTOR kinase when there are enough nutrients and growth factors present. Death receptors such as TNF-R1 and Fas (apoptosis-mediating surface antigen), toll-like receptors, viruses, and DNA damage can trigger necroptosis [14][15].
4. Transcription Factor Nuclear Respiratory Factor 2
Nuclear factor erythroid 2-related factor 2 is a vital transcription factor responsible for regulating antioxidant and detoxifying enzymes through a domain referred to as ARE (antioxidant response element) or EpRE (electrophile responsive element) [16]. The function of Nrf2 is controlled by Keap1, a cytosolic repressor protein also referred to as the cytosolic repressor protein Kelch-like ECH-associated protein 1. Initially, it was believed that Keap1 confines Nrf2 in the cytoplasm and releases it under stress conditions, thus allowing Nrf2 to travel to the nucleus and regulate gene expression. However, further research has demonstrated that Nrf2 is inherently unstable and undergoes degradation by ubiquitin. Therefore, it has been proposed that Keap1 actively promotes the ubiquitination of Nrf2, thereby resulting in its breakdown and preventing its translocation to the nucleus. Follow-up experiments have confirmed this hypothesis [17][18]. Nrf2 is made up of seven conserved regions that are present in different species, which are known as Neh1–7 domains. Neh1 acts as a site for small protein heterodimerization and DNA binding. Neh4 and Neh5 are critical areas for Nrf2 post-activation, while Neh3, situated at the C-terminal end, is considered essential. The Neh2 domain’s N-terminal end is linked to Keap1, which regulates Nrf2 negatively by altering its subcellular structure and increasing its degradation rate. Neh6 is pivotal in controlling Nrf2 degradation in the nucleus. Moreover, Neh7, in conjunction with Neh6, facilitates the additional post-translational negative regulation of Nrf2. This regulation is not reliant on redox, unlike the one mediated by Neh2 and Keap1 [19][20][21][22][23]. The Keap1-Nrf2 pathway is a crucial regulator of the antioxidant system. When ROS are present, Keap1’s cysteine thiols may undergo oxidation, which alters its bonding with Nrf2. Consequently, Nrf2 migrates to the nucleus and pairs with small MAF proteins (musculoaponeurotic fibrosarcoma proteins) forming Nrf2-MAF heterodimers. These heterodimers have the ability to recognize and attach to a specific DNA sequence called ARE/EpRE. Once Nrf2 binds to this region, it activates the transcription of more than 250 genes that contain this sequence in their regulatory regions [24][25][26][27]. During the late 1980s, researchers conducted initial investigations into the presence of AREs, with a primary focus on xenobiotic metabolism. Through these studies, it was revealed that specific compounds could induce xenobiotic metabolism by synthesizing phase I and II enzymes.
Other compounds, on the other hand, only synthesize phase II enzymes, such as glutathione S-transferase (GST), NADPH quinone oxidoreductase (NQO), etc. [28]. The activation of Nrf2 can also occur through the phosphorylation of specific serine and/or tyrosine residues on Keap1. This leads to the separation of Nrf2 and its transfer to the nucleus, where it binds to MAF and ARE, and regulates the expression of protective molecules. These include antioxidant enzymes such as SOD, GPx, CAT, GR, GCL, Trx, TrxR, and PRDX1; detoxification enzymes such as HO-1, NQO1, and GST; GSH-related proteins like γ-GCS (γ-glutamylcysteine synthetase); NADPH-producing enzymes; and various others. By controlling the expression of these proteins, Nrf2 helps to prevent the damage to the body caused by oxidative stress and inflammation (Figure 3) [5].
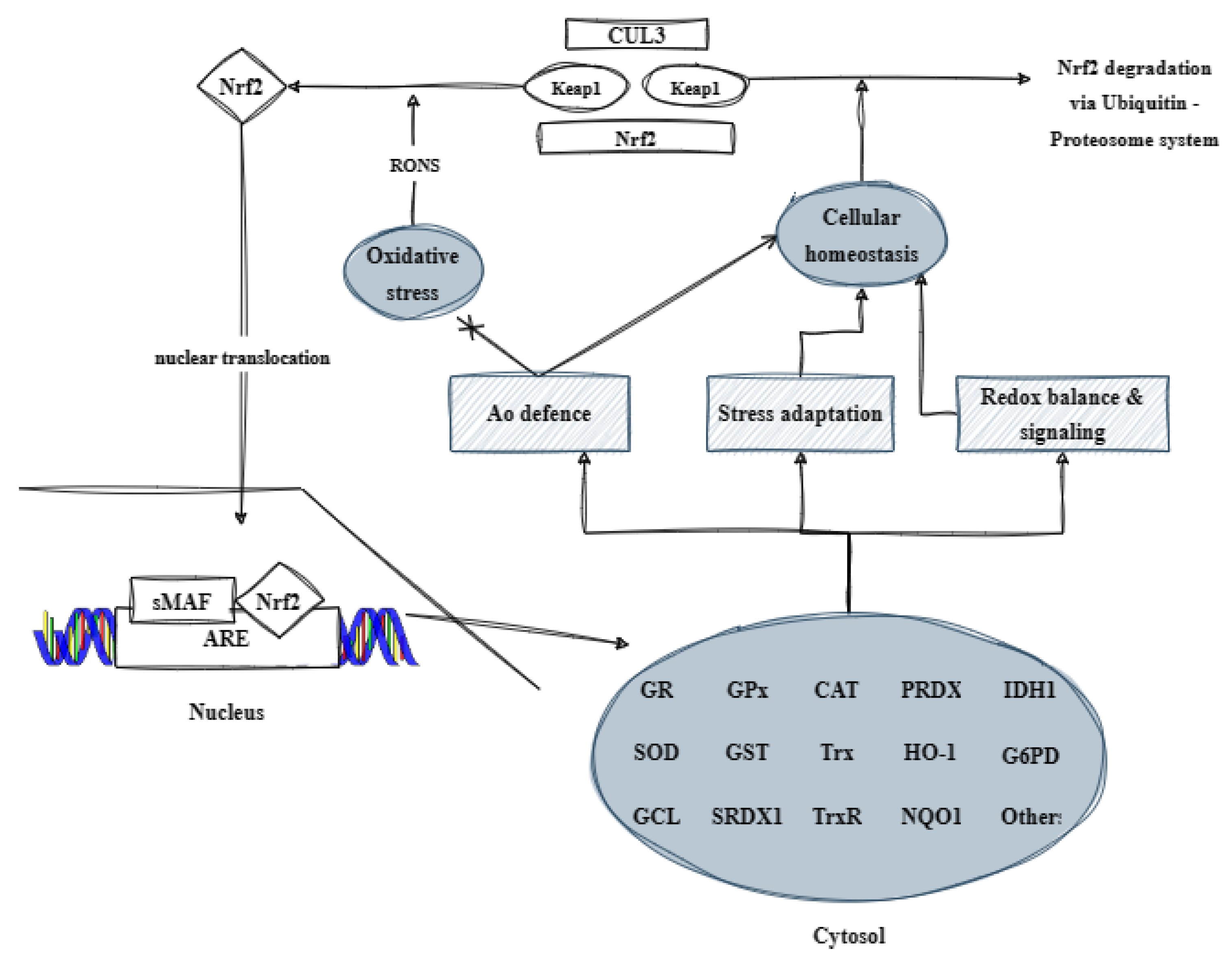
Figure 3. Illustration of the contribution of the transcription factor Nrf2 to the organism’s antioxidant defense (based on [5] with modifications). CUL3: Cullin 3; Keap1: kelch-like ECH-associated protein 1; RONS: reactive oxygen and nitrogen species; Nrf2: nuclear respiratory factor 2; Ao: antioxidant; sMAF: small musculoaponeurotic fibrosarcoma protein; ARE: antioxidant response element; GR: glutathione reductase; GPx: glutathione peroxidase; CAT: catalase; PRDX: peroxiredoxin; IDH1: histidinol dehydrogenase 1; SOD: superoxide dismutase; GST: glutathione S-transferase; Trx: thioredoxin; HO-1: heme oxygenase; G6PD: glucose-6-phosphate dehydrogenase; GCL: glutamate cysteine ligase; SRDX1: EAR motif-based artificial transcriptional repression domain 1; TrxR: thioredoxin reductase; and NQO1: NAD(P)H quinone dehydrogenase 1.
Furthermore, it has been reported that Nrf2 also regulates the expression of the genes that play a crucial role in lipid synthesis and uptake. These genes include SREBPs (sterol regulatory element-binding proteins), FAS (fatty acid synthase), SCD (stearoyl-CoA desaturase), and PPAR (peroxisome proliferator-activated receptor) [29].
4. Transcription Factor Nuclear Factor Kappa B
Nuclear factor kappa B is a transcription factor that can be activated in lymphocytes and plays a crucial role in the body’s inflammatory response. The inflammatory response is a natural process that involves the migration of blood cells and proteins to the site of injury, infection, trauma, or other stimuli that can initiate an immune response. This process is closely linked to oxidative stress. Tissue damage triggers the production of prostaglandins (PGs), leukotrienes (LTs), interleukins (ILs), cytokines, hydrolytic enzymes, and ROS [30]. This response can be triggered by various factors, such as oxidative stress, TNF-α, bacterial lipopolysaccharide (LPS), viruses, UV, and γ-rays, and it is known to regulate the expression of various genes, including pro-inflammatory cytokines, leukocyte adhesion molecules, acute phase proteins, and antimicrobial peptides. Within mammals and birds, the NF-κB protein complex is composed of five subunits, each of which possess the capacity to bind to DNA. These are p50 (or NF-κB1), p52 (or NF-κB2), p65 (or RelA), c-Rel, and RelB, which are encoded by the genes nfkb1, nfkb2, rela, crel, and relb, respectively. The NF-κB family comprises numerous transcription factors possessing a Rel homology domain (RHD) at their N-terminal end, which enables the formation of homo- and heterodimers. The NF-κB dimers are capable of binding to specific DNA regions known as “κB sites” to regulate gene expression. These sites are present in the enhancers and promoters of several genes. In addition, p65, c-Rel, and RelB possess a transcription activation domain (TAD) at their C-terminal end, which aids in the positive regulation of gene expression. On the other hand, subunits that lack the TAD domain (i.e., p50 and p52) can activate transcription by forming heterodimers with p65, c-Rel, or RelB, or by enlisting other proteins with TAD. Homodimers are unable to elicit transcription via DNA binding and instead inhibit the transcription process. The regulatory mechanisms for NF-κB exhibit similarities with those of Nrf2. For instance, the protein exists in an inactive state in the cytoplasm and is bound by inhibitory proteins known as IκB (e.g., IκBα, IκBβ, IκBγ, IκBδ, IκBε, IκBζ, p100, p105, Bcl-3, and IκBns). Under oxidative stress, these two factors compete to coordinate an appropriate response. In the presence of cytokines and other stressors, the activation of NF-κB transpires. Specifically, when a dimer of NF-κB enters the nucleus, the IκB bound to it subsequently undergoes phosphorylation at specific serine residues through the IκB kinase (IKK) complex. The phosphorylation process is a significant event that initiates the NF-κB signaling pathway. This, in turn, leads to the expression of a diverse set of genes that play a crucial role in immune and inflammatory responses. The IKK complex is made up of two active kinases, namely IKKα (IKK1) and IKKβ (IKK2), along with a regulatory scaffolding protein known as NEMO (IKKγ) [31][32][33][34]. Activators of the IKK complex include mitogen-activated protein kinases (MAP3Ks) such as MEKK1, MEKK3, and TAK1 [35]. NF-κB and Nrf2 have an antagonistic relationship, where one inhibits the activation of the other. Nrf2 can prevent the activation of NF-κB by increasing the antioxidant defense, thereby reducing ROS and preventing the degradation of IκBα, as well as inhibiting the transcription of proinflammatory genes. Similarly, Keap1 can also act as IKKβ and negatively regulate NF-κB by stabilizing IκBα, thereby leading to the inhibition of its activation. In contrast, NF-κB may impede the activation of Nrf2 by inducing the recruitment of a histone to the ARE domain, which—in turn—restrains the transcription of the corresponding genes. Nrf2 and p65 compete for the CBP-p300 complex, which is responsible for transferring an acetyl unit to the lysine residues of transcription factors. This enhances gene transcription. Generally, CBP (CREB binding protein) exhibits a preference for κB genes [5][36][37].
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818.
- FAO; IFAD; UNICEF; WFP; WHO. The State of Food Security and Nutrition in the World 2021. Transforming Food Systems for Food Security, Improved Nutrition and Affordable Healthy Diets for All; FAO: Rome, Italy, 2021.
- Korver, D.R. Review: Current challenges in poultry nutrition, health, and welfare. Animal 2023, 17 (Suppl. 2), 100755.
- Poultry Global Market Report. 2023. Available online: https://www.researchandmarkets.com/report/poultry (accessed on 28 January 2024).
- Surai, F.P. Vitagenes in Avian Biology and Poultry Health; Wageningen Academic: Leiden, The Netherlands, 2020; pp. 25–544.
- Surai, P.F. Integrated antioxidant defence network in animals. EC Nutr. 2023, 18, 18–20.
- Surai, P.; Kochish, I.I.; Fisinin, I.V.; Kidd, M.T. Antioxidant defence systems and oxidative stress in poultry biology: An update. Antioxidants 2019, 8, 235.
- George, S.; Abrahamse, H. Redox Potential of Antioxidants in Cancer Progression and Prevention. Antioxidants 2020, 9, 1156.
- Behl, T.; Rana, T.; Alotaibi, G.H.; Shamsuzzaman, M.; Naqvi, M.; Sehgal, A.; Singh, S.; Sharma, N.; Almoshari, Y.; Abdellatif, A.A.H.; et al. Polyphenols inhibiting MAPK signaling pathway mediated oxidative stress and inflammation in depression. Biomed. Pharmacother. 2022, 146, 112545.
- Martindale, J.L.; Holbrook, N.J. Cellular response to oxidative stress: Signaling for suicide and survival. J. Cell Physiol. 2002, 192, 1–15.
- Knasmüller, S.; Nersesyan, A.; Mišík, M.; Gerner, C.; Mikulits, W.; Ehrlich, V.; Hoelzl, C.; Szakmary, A.; Wagner, K.-H. Use of conventional and -omics based methods for health claims of dietary antioxidants: A critical overview. Br. J. Nutr. 2008, 99, ES3–ES52.
- Zoidis, E.; Seremelis, I.; Kontopoulos, N.; Danezis, G.P. Selenium-dependent antioxidant enzymes: Actions and properties of selenoproteins. Antioxidants 2018, 7, 66.
- Surai, P.F.; Fisinin, V.I. Vitagenes in poultry production: Part 3. Vitagene concept development. World’s Poult. Sci. J. 2016, 72, 793–804.
- Iside, C.; Scafuro, M.; Nebbioso, A.; Altucci, L. SIRT1 activation by natural phytochemicals: An overview. Front. Pharmacol. 2020, 11, 1225.
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2977–2992.
- Ishii, T.; Itoh, K.; Takahashi, S.; Sato, H.; Yanagawa, T.; Katoh, Y.; Bannai, S.; Yamamoto, M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000, 275, 16023–16029.
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86.
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-Antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295.
- Itoh, K.; Ishii, T.; Wakabayashi, N.; Yamamoto, M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 1999, 31, 319–324.
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600.
- Hur, W.; Sun, Z.; Jiang, T.; Mason, D.E.; Peters, E.C.; Zhang, D.D.; Luesch, H.; Schultz, P.G.; Gray, N.S. A small-molecule inducer of the antioxidant response element. Chem. Biol. 2010, 17, 537–547.
- Itoh, K.; Mimura, J.; Yamamoto, M. Discovery of the negative regulator of Nrf2, Keap1: A historical overview. Antioxid. Redox Signal. 2010, 13, 1665–1678.
- Zenkov, N.K.; Kozhin, P.M.; Chechushkov, A.V.; Martinovich, G.G.; Kandalintseva, N.V.; Menshchikova, E.B. Mazes of Nrf2 regulation. Biochemistry 2017, 82, 556–564.
- Itoh, K.; Tong, K.I.; Yamamoto, M. Molecular mechanism activating nrf2–keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 2004, 36, 1208–1213.
- Huang, M.L.-H.; Chiang, S.; Kalinowski, D.S.; Bae, D.-H.; Sahni, S.; Richardson, D.R. The role of the antioxidant response in mitochondrial dysfunction in degenerative diseases: Crosstalk between antioxidant defense, autophagy, and apoptosis. Oxid. Med. Cell. Longev. 2019, 2019, 6392763.
- Vargas-Mendoza, N.; Morales-González, Á.; Madrigal-Santillán, E.O.; Madrigal-Bujaidar, E.; Álvarez-González, I.; García-Melo, L.F.; Anguiano-Robledo, L.; Fregoso-Aguilar, T.; Morales-Gonzalez, J.A. Antioxidant and adaptative response mediated by Nrf2 during physical exercise. Antioxidants 2019, 8, 196.
- Hannan, M.A.; Dash, R.; Sohag, A.A.M.; Haque, M.N.; Moon, I.S. Neuroprotection Against Oxidative Stress: Phytochemicals Targeting TrkB Signaling and the Nrf2-ARE Antioxidant System. Front. Mol. Neurosci. 2020, 13, 116.
- Lyakhovich, V.V.; Vavilin, V.A.; Zenkov, N.K.; Menshchikova, E.B. Active defense under oxidative stress. The antioxidant responsive element. Biochemistry 2006, 71, 962–974.
- Chambel, S.S.; Santos-Gonçalves, A.; Duarte, T.L. The dual role of Nrf2 in nonalcoholic fatty liver disease: Regulation of Antioxidant Defenses and Hepatic Lipid Metabolism. BioMed Res. Int. 2015, 2015, 597134.
- Sagin, F.; Sozmen, E. Anti-inflammatory effects of dietary antioxidants. Antiinflamm. Antiallergy Agents Med. Chem. 2004, 3, 19–30.
- Shen, G.; Jeong, W.-S.; Hu, R.; Kong, A.-N.T. Regulation of Nrf2, NF-κB, and AP-1 signaling pathways by chemopreventive agents. Antioxid. Redox Signal. 2005, 7, 1648–1663.
- O’Dea, E.; Hoffmann, A. NF-κB signaling. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 107–115.
- Hayden, S.M.; Ghosh, S. NF-κB in immunobiology. Cell Res. 2011, 21, 223–244.
- Surai, P.F.; Kochish, I.I.; Kidd, M.T. Redox homeostasis in poultry: Regulatory roles of NF-κB. Antioxidants 2021, 10, 186.
- Moynagh, P.N. The NF-B pathway. J. Cell Sci. 2005, 118, 4589–4592.
- Lingappan, K. NF-κB in oxidative stress. Curr. Opin. Toxicol. 2018, 7, 81–86.
- Gado, F.; Ferrario, G.; Della Vedova, L.; Zoanni, B.; Altomare, A.; Carini, M.; Aldini, G.; D’Amato, A.; Baron, G. Targeting Nrf2 and NF-κB signaling pathways in cancer prevention: The role of apple phytochemicals. Molecules 2023, 28, 1356.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
789
Revisions:
2 times
(View History)
Update Date:
15 Apr 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




